6JW4
 
 | | Degenerate RVD RG forms a distinct loop conformation | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5HC)P*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | Authors: | Liu, L, Yi, C. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
6JW5
 
 | | RVD Q* recognizes 5hmC through water-mediated H bonds | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5HC)P*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | Authors: | Liu, L, Yi, C. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
6JW0
 
 | | Universal RVD R* accommodates cytosine via water-mediated interactions | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*CP*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | Authors: | Liu, L, Yi, C. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
6JW3
 
 | | Degenerate RVD RG forms a distinct loop conformation | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5CM)P*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | Authors: | Liu, L, Yi, C. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
6JW1
 
 | | Universal RVD R* accommodates 5mC via water-mediated interactions | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5CM)P*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | Authors: | Liu, L, Yi, C. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
6JW2
 
 | | Universal RVD R* accommodates 5hmC via water-mediated interactions | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5HC)P*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | Authors: | Liu, L, Yi, C. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
6JVZ
 
 | | RVD HA specifically contacts 5mC through van der Waals interactions | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5CM)P*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | Authors: | Liu, L, Yi, C. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
5I2K
 
 | | Structure of the human GluN1/GluN2A LBD in complex with 7-{[ethyl(4-fluorophenyl)amino]methyl}-N,2-dimethyl-5-oxo-5H-[1,3]thiazolo[3,2-a]pyrimidine-3-carboxamide (compound 19) | | Descriptor: | 7-{[ethyl(4-fluorophenyl)amino]methyl}-N,2-dimethyl-5-oxo-5H-[1,3]thiazolo[3,2-a]pyrimidine-3-carboxamide, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-02-09 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Discovery of GluN2A-Selective NMDA Receptor Positive Allosteric Modulators (PAMs): Tuning Deactivation Kinetics via Structure-Based Design.
J.Med.Chem., 59, 2016
|
|
5I2N
 
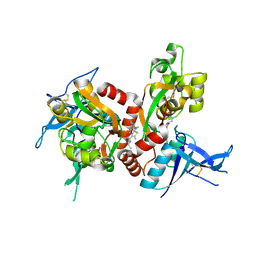 | | Structure of the human GluN1/GluN2A LBD in complex with N-ethyl-7-{[2-fluoro-3-(trifluoromethyl)phenyl]methyl}-2-methyl-5-oxo-5H-[1,3]thiazolo[3,2-a]pyrimidine-3-carboxamide (compound 29) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, GLUTAMIC ACID, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-02-09 | | Release date: | 2016-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Discovery of GluN2A-Selective NMDA Receptor Positive Allosteric Modulators (PAMs): Tuning Deactivation Kinetics via Structure-Based Design.
J.Med.Chem., 59, 2016
|
|
5KDT
 
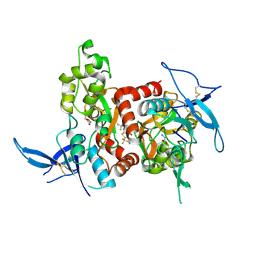 | | Structure of the human GluN1/GluN2A LBD in complex with GNE0723 | | Descriptor: | (1~{R},2~{R})-2-[7-[[5-chloranyl-3-(trifluoromethyl)pyrazol-1-yl]methyl]-5-oxidanylidene-2-(trifluoromethyl)-[1,3]thiazolo[3,2-a]pyrimidin-3-yl]cyclopropane-1-carbonitrile, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-06-08 | | Release date: | 2016-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Discovery of GluN2A-Selective NMDA Receptor Positive Allosteric Modulators (PAMs): Tuning Deactivation Kinetics via Structure-Based Design.
J. Med. Chem., 59, 2016
|
|
6JQ4
 
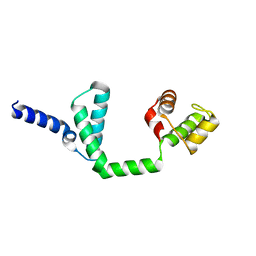 | | HIGA Escherichia coli-K12 | | Descriptor: | Antitoxin HigA | | Authors: | She, Z, Xu, B.S. | | Deposit date: | 2019-03-28 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes of antitoxin HigA from Escherichia coli str. K-12 upon binding of its cognate toxin HigB reveal a new regulation mechanism in toxin-antitoxin systems.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
7C02
 
 | | Crystal structure of dimeric MERS-CoV receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Dai, L, Qi, J, Gao, G.F. | | Deposit date: | 2020-04-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A Universal Design of Betacoronavirus Vaccines against COVID-19, MERS, and SARS.
Cell, 182, 2020
|
|
6O8B
 
 | | Crystal structure of STING CTD in complex with TBK1 | | Descriptor: | N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1, Stimulator of interferon genes protein | | Authors: | Li, P, Zhao, B, Du, F. | | Deposit date: | 2019-03-09 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A conserved PLPLRT/SD motif of STING mediates the recruitment and activation of TBK1.
Nature, 569, 2019
|
|
6O8C
 
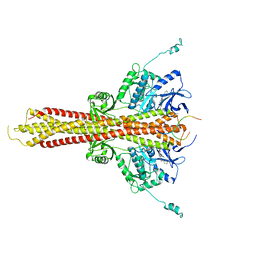 | | Crystal structure of STING CTT in complex with TBK1 | | Descriptor: | N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1, Stimulator of interferon genes protein | | Authors: | Li, P, Zhao, B, Du, F. | | Deposit date: | 2019-03-09 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | A conserved PLPLRT/SD motif of STING mediates the recruitment and activation of TBK1.
Nature, 569, 2019
|
|
6LU7
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor N3 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | Deposit date: | 2020-01-26 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
4R99
 
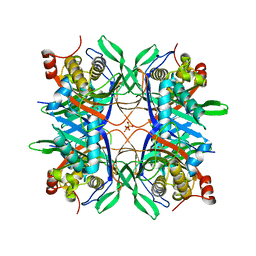 | | Crystal structure of a uricase from Bacillus fastidious | | Descriptor: | SULFATE ION, Uricase | | Authors: | Feng, J, Wang, L, Liu, H.B, Liu, L, Liao, F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bacillus fastidious uricase reveals an unexpected folding of the C-terminus residues crucial for thermostability under physiological conditions.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4R8X
 
 | | Crystal structure of a uricase from Bacillus fastidious | | Descriptor: | Uricase | | Authors: | Feng, J, Wang, L, Liu, H.B, Liu, L, Liao, F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Crystal structure of Bacillus fastidious uricase reveals an unexpected folding of the C-terminus residues crucial for thermostability under physiological conditions.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
2H96
 
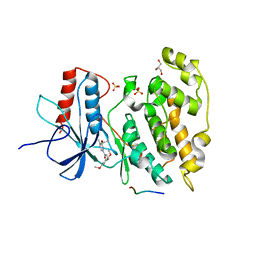 | | Discovery of Potent, Highly Selective, and Orally Bioavailable Pyridine Carboxamide C-jun NH2-terminal Kinase Inhibitors | | Descriptor: | 5-CYANO-N-(2,5-DIMETHOXYBENZYL)-6-ETHOXYPYRIDINE-2-CARBOXAMIDE, C-jun-amino-terminal kinase-interacting protein 1, GLYCEROL, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2006-06-09 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of potent, highly selective, and orally bioavailable pyridine carboxamide c-Jun NH2-terminal kinase inhibitors.
J.Med.Chem., 49, 2006
|
|
7WKJ
 
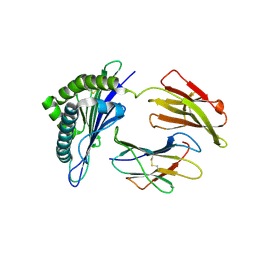 | | A COVID-19 T-cell response detection method based on a newly identified human CD8+ T cell epitope from SARS-CoV-2-Hubei Province, 2021. | | Descriptor: | Beta-2-microglobulin, LYS-THR-PHE-PRO-PRO-THR-GLU-PRO-LYS, MHC class I antigen | | Authors: | Zhang, J, Lu, D, Li, M, Liu, M.S, Yao, S.J, Zhan, J.B, Liu, J, Gao, G.F. | | Deposit date: | 2022-01-10 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A COVID-19 T-Cell Response Detection Method Based on a Newly Identified Human CD8 + T Cell Epitope from SARS-CoV-2 - Hubei Province, China, 2021.
China CDC Wkly, 4, 2022
|
|
5AY7
 
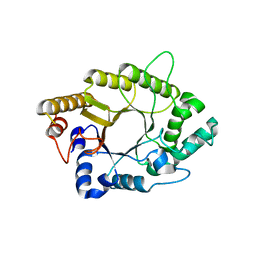 | | A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase | | Descriptor: | xylanase | | Authors: | Zheng, Y, Li, Y, Liu, W, Guo, R.T. | | Deposit date: | 2015-08-10 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insight into potential cold adaptation mechanism through a psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase.
J.Struct.Biol., 193, 2016
|
|
5UHT
 
 | | Structure of the Thermotoga maritima HK853-BeF3-RR468 complex at pH 5.0 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Liu, Y, Rose, J, Jiang, L, Zhou, P. | | Deposit date: | 2017-01-12 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | A pH-gated conformational switch regulates the phosphatase activity of bifunctional HisKA-family histidine kinases.
Nat Commun, 8, 2017
|
|
8IEX
 
 | | Solution structure of AtWRKY11-DBD | | Descriptor: | Probable WRKY transcription factor 11, ZINC ION | | Authors: | Dong, X, Hu, Y.F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-25 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of Arabidopsis transcription factor WRKY11.
Biochem.Biophys.Res.Commun., 653, 2023
|
|
8IF5
 
 | | AFB1-AF26 APTAMER COMPLEX | | Descriptor: | AFB1 DNA aptamer (26-MER), AFLATOXIN B1 | | Authors: | Xu, G.H, Wang, C, Li, C.G. | | Deposit date: | 2023-02-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for high-affinity recognition of aflatoxin B1 by a DNA aptamer.
Nucleic Acids Res., 51, 2023
|
|
4DDP
 
 | |
4ZP0
 
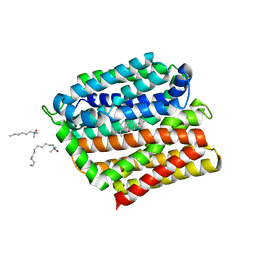 | | Crystal structure of E. coli multidrug transporter MdfA in complex with deoxycholate | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, LAURYL DIMETHYLAMINE-N-OXIDE, Multidrug transporter MdfA | | Authors: | Zhang, X.C, Heng, J, Zhao, Y, Wang, X. | | Deposit date: | 2015-05-07 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate-bound structure of the E. coli multidrug resistance transporter MdfA
Cell Res., 25, 2015
|
|
