6KMO
 
 | |
7CN4
 
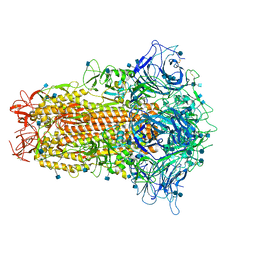 | | Cryo-EM structure of bat RaTG13 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Zhang, S, Qiao, S, Yu, J, Zeng, J, Tian, L. | | Deposit date: | 2020-07-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution.
Nat Commun, 12, 2021
|
|
7CN8
 
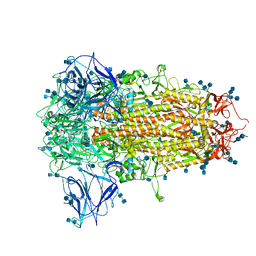 | | Cryo-EM structure of PCoV_GX spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein, ... | | Authors: | Wang, X, Yu, J, Zhang, S, Qiao, S, Zeng, J, Tian, L. | | Deposit date: | 2020-07-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution.
Nat Commun, 12, 2021
|
|
7FAE
 
 | | S protein of SARS-CoV-2 in complex bound with P36-5D2(state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, P36-5D2 heavy chain, ... | | Authors: | Zhang, L, Wang, X, Shan, S, Zhang, S. | | Deposit date: | 2021-07-06 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | A Potent and Protective Human Neutralizing Antibody Against SARS-CoV-2 Variants.
Front Immunol, 12, 2021
|
|
7FAF
 
 | | S protein of SARS-CoV-2 in complex bound with P36-5D2 (state1) | | Descriptor: | P36-5D2 heavy chain, P36-5D2 light chain, Spike glycoprotein | | Authors: | Zhang, L, Wang, X, Zhang, S, Shan, S. | | Deposit date: | 2021-07-06 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | A Potent and Protective Human Neutralizing Antibody Against SARS-CoV-2 Variants.
Front Immunol, 12, 2021
|
|
7F7E
 
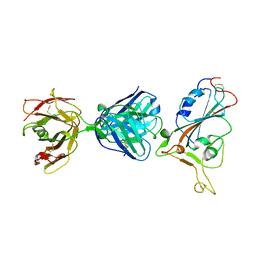 | | SARS-CoV-2 S protein RBD in complex with A5-10 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of A5-10 Fab, Light chain of A5-10 Fab, ... | | Authors: | Dou, Y, Wang, X, Wang, K, Liu, P, Lu, B. | | Deposit date: | 2021-06-29 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Etesevimab in combination with JS026 neutralizing SARS-CoV-2 and its variants.
Emerg Microbes Infect, 11, 2022
|
|
5D9L
 
 | | Rsk2 N-terminal Kinase in Complex with bis-phenol pyrazole | | Descriptor: | 4,4'-(1H-pyrazole-3,4-diyl)diphenol, GLYCEROL, Ribosomal protein S6 kinase alpha-3 | | Authors: | Appleton, B.A. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of Potent and Selective RSK Inhibitors as Biological Probes.
J.Med.Chem., 58, 2015
|
|
7C8V
 
 | | Structure of sybody SR4 in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7C8W
 
 | | Structure of sybody MR17 in complex with the SARS-CoV-2 S receptor-binding domain (RBD) | | Descriptor: | GLYCEROL, Spike protein S1, Synthetic nanobody MR17, ... | | Authors: | Li, T, Cai, H, Yao, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7CAN
 
 | | Structure of sybody MR17-K99Y in complex with the SARS-CoV-2 S Receptor-binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-09 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
5GSB
 
 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 37-3 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-D alpha chain, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-15 | | Release date: | 2017-07-12 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
6J11
 
 | | MERS-CoV spike N-terminal domain and 7D10 scFv complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-terminal domain of Spike glycoprotein, ... | | Authors: | Zhou, H, Zhang, S, Zhang, S, Tang, W, Wang, X. | | Deposit date: | 2018-12-27 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural definition of a neutralization epitope on the N-terminal domain of MERS-CoV spike glycoprotein.
Nat Commun, 10, 2019
|
|
7B3N
 
 | | AmiP amidase-3 from Thermus parvatiensis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Cell wall hydrolase, ... | | Authors: | Freitag-Pohl, S, Pohl, E. | | Deposit date: | 2020-12-01 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | AmiP from hyperthermophilic Thermus parvatiensis prophage is a thermoactive and ultrathermostable peptidoglycan lytic amidase.
Protein Sci., 32, 2023
|
|
4N70
 
 | | Pim1 Complexed with a pyridylcarboxamide | | Descriptor: | N-{4-[(3R,4R,5S)-3-amino-4-hydroxy-5-methylpiperidin-1-yl]pyridin-3-yl}-6-(2,6-difluorophenyl)-5-fluoropyridine-2-carboxamide, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C.R, Le, V, Shu, W, Burger, M.T, Bussiere, D. | | Deposit date: | 2013-10-14 | | Release date: | 2013-11-06 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure Guided Optimization, in Vitro Activity, and in Vivo Activity of Pan-PIM Kinase Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
5YAN
 
 | |
4N6Y
 
 | | Pim1 Complexed with a phenylcarboxamide | | Descriptor: | 2-(acetylamino)-N-[2-(piperidin-1-yl)phenyl]-1,3-thiazole-4-carboxamide, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C.R, Le, V, Shu, W, Burger, M.T, Bussiere, D. | | Deposit date: | 2013-10-14 | | Release date: | 2013-11-06 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure Guided Optimization, in Vitro Activity, and in Vivo Activity of Pan-PIM Kinase Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
6K5E
 
 | | Crystal structure of BioH from Klebsiella pneumonia | | Descriptor: | Pimeloyl-[acyl-carrier protein] methyl ester esterase | | Authors: | Wang, L, Chen, Y. | | Deposit date: | 2019-05-28 | | Release date: | 2019-07-17 | | Last modified: | 2020-05-06 | | Method: | X-RAY DIFFRACTION (2.257 Å) | | Cite: | Structural insight into the carboxylesterase BioH from Klebsiella pneumoniae.
Biochem.Biophys.Res.Commun., 520, 2019
|
|
4N6Z
 
 | | Pim1 Complexed with a pyridylcarboxamide | | Descriptor: | 3-amino-N-{4-[(3S)-3-aminopiperidin-1-yl]pyridin-3-yl}pyrazine-2-carboxamide, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C.R, Le, V, Shu, W, Burger, M.T, Bussiere, D. | | Deposit date: | 2013-10-14 | | Release date: | 2013-11-06 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Guided Optimization, in Vitro Activity, and in Vivo Activity of Pan-PIM Kinase Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
7FDK
 
 | | SARS-COV-2 Spike RBDMACSp36 binding to mACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7FDH
 
 | | SARS-COV-2 Spike RBDMACSp25 binding to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7FDI
 
 | | SARS-COV-2 Spike RBDMACSp36 binding to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7FDG
 
 | | SARS-COV-2 Spike RBDMACSp6 binding to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7DMW
 
 | | Crystal structure of CcpC regulatory domain in complex with citrate from Bacillus amyloliquefaciens | | Descriptor: | CITRATE ANION, CcpC | | Authors: | Chen, J, Wang, L, Shang, F, Liu, W, Chen, Y, Lan, J, Bu, T, Bai, X, Xu, Y. | | Deposit date: | 2020-12-08 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Functional and structural analysis of catabolite control protein C that responds to citrate.
Sci Rep, 11, 2021
|
|
5D1Z
 
 | | IsdB NEAT1 bound by clone D4-10 | | Descriptor: | D4-10 Heavy Chain, D4-10 Light Chain, Iron-regulated surface determinant protein B, ... | | Authors: | Deng, X. | | Deposit date: | 2015-08-04 | | Release date: | 2016-09-14 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Germline-encoded neutralization of a Staphylococcus aureus virulence factor by the human antibody repertoire.
Nat Commun, 7, 2016
|
|
5D1Q
 
 | | IsdB NEAT2 bound by clone D2-06 | | Descriptor: | D2-06 Heavy Chain, D2-06 Light Chain, Iron-regulated surface determinant protein B, ... | | Authors: | Deng, X. | | Deposit date: | 2015-08-04 | | Release date: | 2016-08-10 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Germline-encoded neutralization of a Staphylococcus aureus virulence factor by the human antibody repertoire.
Nat Commun, 7, 2016
|
|
