2J14
 
 | | 3,4,5-Trisubstituted Isoxazoles as Novel PPARdelta Agonists: Part2 | | Descriptor: | (3-{4-[2-(2,4-DICHLORO-PHENOXY)-ETHYLCARBAMOYL]-5-PHENYL-ISOXAZOL-3-YL}-PHENYL)-ACETIC ACID, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR DELTA | | Authors: | Epple, R, Azimioara, M, Russo, R, Xie, Y, Wang, X, Cow, C, Wityak, J, Karanewsky, D, Bursulaya, B, Kreusch, A, Tuntland, T, Gerken, A, Iskandar, M, Saez, E, Seidel, H.M, Tian, S.S. | | Deposit date: | 2006-08-08 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 3,4,5-Trisubstituted Isoxazoles as Novel Ppardelta Agonists. Part 2
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4XTB
 
 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter | | Descriptor: | Calcium uniporter protein, mitochondrial, TETRAETHYLENE GLYCOL | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-01-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
1WP1
 
 | | Crystal structure of the drug-discharge outer membrane protein, OprM | | Descriptor: | Outer membrane protein oprM | | Authors: | Akama, H, Kanemaki, M, Yoshimura, M, Tsukihara, T, Kashiwagi, T, Narita, S, Nakagawa, A, Nakae, T. | | Deposit date: | 2004-08-28 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end
J.Biol.Chem., 279, 2004
|
|
8SEF
 
 | | Crystal structure of Cy137C02, a monoclonal antibody isolated from macaques immunized with an Epstein-Barr virus glycoprotein 350 (gp350) nanoparticle vaccine | | Descriptor: | Cy137C02 Fab heavy chain, Cy137C02 Fab light chain, GLYCEROL, ... | | Authors: | Joyce, M.G, Jensen, J.L, Chen, W.H, Kanekiyo, M. | | Deposit date: | 2023-04-09 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of Cy137C02, a monoclonal antibody isolated from macaques immunized with an Epstein-Barr virus glycoprotein 350 (gp350) nanoparticle vaccine
To Be Published
|
|
8SGN
 
 | | Crystal structure of Epstein-Barr virus glycoprotein 350 (gp350) in complex with Cy651H02, a monoclonal antibody isolated from macaques immunized with a gp350 nanoparticle vaccine | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Joyce, M.G, Jensen, J.L, Chen, W.H, Kanekiyo, M. | | Deposit date: | 2023-04-12 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Epstein-Barr virus glycoprotein 350 (gp350) in complex with Cy651H02, a monoclonal antibody isolated from macaques immunized with a gp350 nanoparticle vaccine
To Be Published
|
|
8SIC
 
 | | Crystal structure of Epstein-Barr virus glycoprotein 350 (gp350) in complex with Cy137C02, a monoclonal antibody isolated from macaques immunized with a gp350 nanoparticle vaccine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cy137C02 Fab heavy chain, Cy137C02 Fab light chain, ... | | Authors: | Joyce, M.G, Jensen, J.L, Chen, W.H, Kanekiyo, M. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Crystal structure of Epstein-Barr virus glycoprotein 350 (gp350) in complex with Cy137C02, a monoclonal antibody isolated from macaques immunized with a gp350 nanoparticle vaccine
To Be Published
|
|
8SGG
 
 | | Crystal structure of Cy137D09, a monoclonal antibody isolated from macaques immunized with an Epstein-Barr virus glycoprotein 350 (gp350) nanoparticle vaccine | | Descriptor: | Cy137D09 Fab heavy chain, Cy137D09 Fab light chain, GLYCEROL | | Authors: | Joyce, M.G, Jensen, J.L, Chen, W.H, Kanekiyo, M. | | Deposit date: | 2023-04-12 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Cy137D09, a monoclonal antibody isolated from macaques immunized with an Epstein-Barr virus glycoprotein 350 (gp350) nanoparticle vaccine
To Be Published
|
|
8SM1
 
 | | CRYSTAL STRUCTURE OF HUMAN ANTIBODY 769A9 IN COMPLEX WITH EPSTEIN-BARR VIRUS MAJOR GLYCOPROTEIN GP350 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769A9 Fab heavy chain, 769A9 Fab light chain, ... | | Authors: | Chen, W.-H, Bu, W, Cohen, J.I, Kanekiyo, M, Joyce, M.G. | | Deposit date: | 2023-04-25 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural Basis For Receptor Engagement And Virus Neutralization Through Epstein-Barr Virus Gp350
To Be Published
|
|
8SM0
 
 | | Crystal structure of human complement receptor 2 (CD21) in complex with Epstein-Barr virus major glycoprotein gp350 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement receptor type 2, Envelope glycoprotein gp350, ... | | Authors: | Chen, W.-H, Bu, W, Cohen, J.I, Kanekiyo, M, Joyce, M.G. | | Deposit date: | 2023-04-25 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Basis For Receptor Engagement And Virus Neutralization Through Epstein-Barr Virus Gp350
To Be Published
|
|
5AWE
 
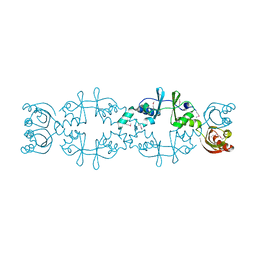 | | Crystal structure of a hypothetical protein, TTHA0829 from Thermus thermophilus HB8, composed of cystathionine-beta-synthase (CBS) and aspartate-kinase chorismate-mutase tyrA (ACT) domains | | Descriptor: | Putative acetoin utilization protein, acetoin dehydrogenase | | Authors: | Nakabayashi, M, Shibata, N, Kanagawa, M, Nakagawa, N, Kuramitsu, S, Higuchi, Y. | | Deposit date: | 2015-07-03 | | Release date: | 2016-05-18 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of a hypothetical protein, TTHA0829 from Thermus thermophilus HB8, composed of cystathionine-beta-synthase (CBS) and aspartate-kinase chorismate-mutase tyrA (ACT) domains.
Extremophiles, 20, 2016
|
|
1YWN
 
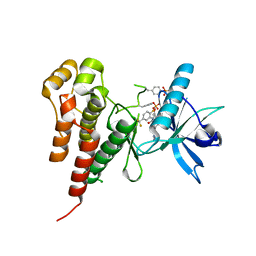 | | Vegfr2 in complex with a novel 4-amino-furo[2,3-d]pyrimidine | | Descriptor: | N-{4-[4-AMINO-6-(4-METHOXYPHENYL)FURO[2,3-D]PYRIMIDIN-5-YL]PHENYL}-N'-[2-FLUORO-5-(TRIFLUOROMETHYL)PHENYL]UREA, Vascular endothelial growth factor receptor 2 | | Authors: | Miyazaki, Y, Matsunaga, S, Tang, J, Maeda, Y, Nakano, M, Philippe, R.J, Shibahara, M, Liu, W, Sato, H, Wang, L, Nolte, R.T. | | Deposit date: | 2005-02-18 | | Release date: | 2005-08-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Novel 4-amino-furo[2,3-d]pyrimidines as Tie-2 and VEGFR2 dual inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2LMK
 
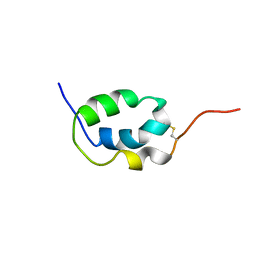 | | Solution Structure of Mouse Pheromone ESP1 | | Descriptor: | Exocrine gland-secreting peptide 1 | | Authors: | Yoshinaga, S, Sato, T, Hirakane, M, Esaki, K, Hamaguchi, T, Haga-Yamanaka, S, Tsunoda, M, Kimoto, H, Shimada, I, Touhara, K, Terasawa, H. | | Deposit date: | 2011-12-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the Mouse Sex Peptide Pheromone ESP1 Reveals a Molecular Basis for Specific Binding to the Class-C G-Protein-Coupled Vomeronasal Receptor
J.Biol.Chem., 2013
|
|
6FZA
 
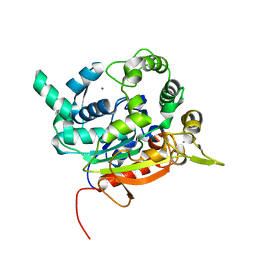 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant A187F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6FZ8
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant L184F/A187F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6FZD
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 variant L184F/A187F/L360F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6FZ7
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant L184F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.736 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6TF6
 
 | | Human galectin-3c in complex with a galactose derivative | | Descriptor: | CHLORIDE ION, Galectin-3, ~{N}-[[(2~{S},3~{S},4~{R},5~{S},6~{R})-4-[[5,6-bis(fluoranyl)-2-oxidanylidene-chromen-3-yl]methoxy]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]methyl]-4-fluoranyl-naphthalene-1-carboxamide | | Authors: | Nilsson, U.J, Zetterberg, F, Hakansson, M, Logan, D.T. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 3-Substituted 1-Naphthamidomethyl-C-galactosyls Interact with Two Unique Sub-sites for High-Affinity and High-Selectivity Inhibition of Galectin-3.
Molecules, 24, 2019
|
|
6FZ1
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant L360F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-13 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
4HD6
 
 | | Crystal Structure of Tyrosinase from Bacillus megaterium V218F mutant soaked in CuSO4 | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Goldfeder, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Influencing the monophenolase/diphenolase activity ratio in tyrosinase.
Biochim.Biophys.Acta, 1834, 2013
|
|
4HD4
 
 | | Crystal Structure of Tyrosinase from Bacillus megaterium V218F mutant | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Goldfeder, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Influencing the monophenolase/diphenolase activity ratio in tyrosinase.
Biochim.Biophys.Acta, 1834, 2013
|
|
4HD7
 
 | | Crystal Structure of Tyrosinase from Bacillus megaterium V218G mutant soaked in CuSO4 | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Goldfeder, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Influencing the monophenolase/diphenolase activity ratio in tyrosinase.
Biochim.Biophys.Acta, 1834, 2013
|
|
6TF7
 
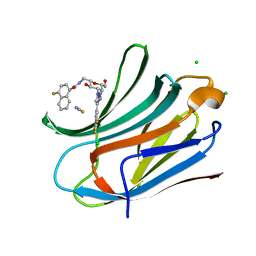 | | Human galectin-3c in complex with a galactose derivative | | Descriptor: | 4-fluoranyl-~{N}-[[(2~{S},3~{R},4~{R},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxan-2-yl]methyl]naphthalene-1-carboxamide, CHLORIDE ION, Galectin-3, ... | | Authors: | Nilsson, U.J, Zetterberg, F, Hakansson, M, Logan, D.T. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 3-Substituted 1-Naphthamidomethyl-C-galactosyls Interact with Two Unique Sub-sites for High-Affinity and High-Selectivity Inhibition of Galectin-3.
Molecules, 24, 2019
|
|
5GTC
 
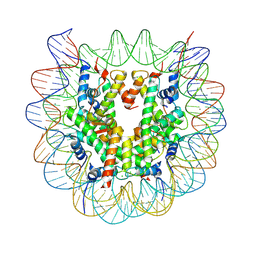 | | Crystal structure of complex between DMAP-SH conjugated with a Kaposi's sarcoma herpesvirus LANA peptide (5-15) and nucleosome core particle | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Arimura, Y, Kato, D, Suto, H, Kurumizaka, H, Kawashima, S.A, Yamatsugu, K, Kanai, M. | | Deposit date: | 2016-08-19 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthetic Posttranslational Modifications: Chemical Catalyst-Driven Regioselective Histone Acylation of Native Chromatin.
J. Am. Chem. Soc., 139, 2017
|
|
8GAT
 
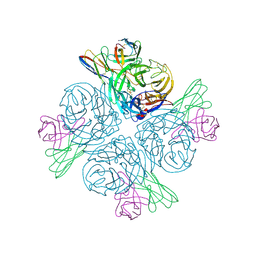 | | Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v), based on consensus cryo-EM map with only Fab 1G01 resolved | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 1G01, heavy chain, ... | | Authors: | Tsybovsky, Y, Lederhofer, J, Kwong, P.D, Kanekiyo, M. | | Deposit date: | 2023-02-23 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Protective human monoclonal antibodies target conserved sites of vulnerability on the underside of influenza virus neuraminidase.
Immunity, 57, 2024
|
|
8GAU
 
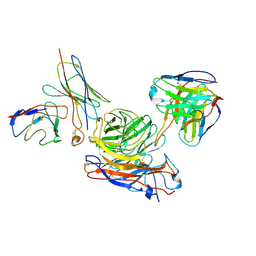 | | Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 1G01, heavy chain, ... | | Authors: | Tsybovsky, Y, Lederhofer, J, Kwong, P.D, Kanekiyo, M. | | Deposit date: | 2023-02-23 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Protective human monoclonal antibodies target conserved sites of vulnerability on the underside of influenza virus neuraminidase.
Immunity, 57, 2024
|
|
