1NR9
 
 | | Crystal Structure of Escherichia coli 1262 (APC5008), Putative Isomerase | | Descriptor: | MAGNESIUM ION, Protein YCGM | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-24 | | Release date: | 2003-07-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Escherichia coli Putative Isomerase EC1262 (APC5008)
To be Published
|
|
3F6V
 
 | | Crystal structure of Possible transcriptional regulator for arsenical resistance | | Descriptor: | MAGNESIUM ION, Possible transcriptional regulator, ArsR family protein | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-06 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of Possible transcriptional regulator for arsenical resistance from Rhodococcus sp.
To be Published
|
|
3FBU
 
 | | The crystal structure of the acetyltransferase (GNAT family) from Bacillus anthracis | | Descriptor: | Acetyltransferase, GNAT family, COENZYME A | | Authors: | Zhang, R, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the acetyltransferase (GNAT family) from Bacillus anthracis
To be Published
|
|
3DCL
 
 | | Crystal structure of TM1086 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SULFATE ION, ... | | Authors: | Chruszcz, M, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-03 | | Release date: | 2008-08-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of TM1086
To be Published
|
|
6OZ1
 
 | | Crystal structure of the adenylation (A) domain of the carboxylate reductase (CAR) GR01_22995 from Mycobacterium chelonae | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Fedorchuk, T, Khusnutdinova, A, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | One-Pot Biocatalytic Transformation of Adipic Acid to 6-Aminocaproic Acid and 1,6-Hexamethylenediamine Using Carboxylic Acid Reductases and Transaminases.
J.Am.Chem.Soc., 142, 2020
|
|
6OX6
 
 | | Crystal structure of the complex between the Type VI effector Tas1 and its immunity protein | | Descriptor: | ACETATE ION, PA14_01140, Tas1 | | Authors: | Ahmad, S, Stogios, P.J, Skarina, T, Whitney, J, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-13 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | An interbacterial toxin inhibits target cell growth by synthesizing (p)ppApp.
Nature, 575, 2019
|
|
6U69
 
 | | Crystal structure of Yck2 from Candida albicans, apoenzyme | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-29 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Overcoming Fungal Echinocandin Resistance through Inhibition of the Non-essential Stress Kinase Yck2.
Cell Chem Biol, 27, 2020
|
|
3DCA
 
 | | Crystal structure of the RPA0582- protein of unknown function from Rhodopseudomonas palustris- a structural genomics target | | Descriptor: | RPA0582, SULFATE ION | | Authors: | Sledz, P, Wang, S, Chruszcz, M, Yim, V, Kudritska, M, Evdokimova, E, Turk, D, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-03 | | Release date: | 2008-08-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal structure of the RPA0582- protein of unknown function from Rhodopseudomonas palustris- a structural genomics target
To be Published
|
|
1MKZ
 
 | | Crystal structure of MoaB protein at 1.6 A resolution. | | Descriptor: | ACETIC ACID, Molybdenum cofactor biosynthesis protein B, SULFATE ION | | Authors: | Sanishvili, R, Skarina, T, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-29 | | Release date: | 2003-04-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Escherichia coli MoaB suggests a probable role in molybdenum cofactor synthesis.
J.Biol.Chem., 279, 2004
|
|
2KKY
 
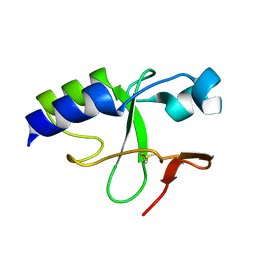 | | Solution Structure of C-terminal domain of oxidized NleG2-3 (residue 90-191) from Pathogenic E. coli O157:H7. Northeast Structural Genomics Consortium and Midwest Center for Structural Genomics target ET109A | | Descriptor: | Uncharacterized protein ECs2156 | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Semest, A, Claude, M, Singer, A, Edwards, A, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-25 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NleG Type 3 effectors from enterohaemorrhagic Escherichia coli are U-Box E3 ubiquitin ligases.
Plos Pathog., 6, 2010
|
|
1INL
 
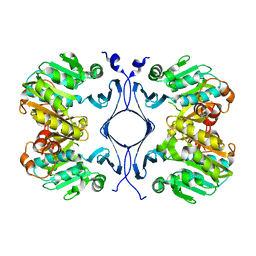 | | Crystal Structure of Spermidine Synthase from Thermotoga Maritima | | Descriptor: | Spermidine synthase | | Authors: | Korolev, S, Skarina, T, Ikeguchi, Y, Pegg, A.E, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-05-14 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of spermidine synthase with a multisubstrate adduct inhibitor.
Nat.Struct.Biol., 9, 2002
|
|
6U6A
 
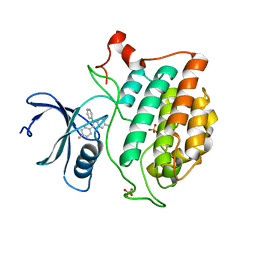 | | Crystal structure of Yck2 from Candida albicans in complex with kinase inhibitor GW461484A | | Descriptor: | 2-(4-fluorophenyl)-6-methyl-3-(pyridin-4-yl)pyrazolo[1,5-a]pyridine, SULFATE ION, Serine/threonine protein kinase | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Chang, C, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-29 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Overcoming Fungal Echinocandin Resistance through Inhibition of the Non-essential Stress Kinase Yck2.
Cell Chem Biol, 27, 2020
|
|
5UVI
 
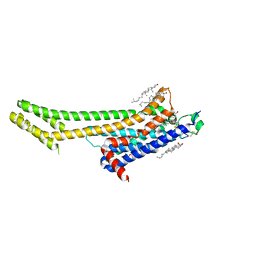 | | Serial Millisecond Crystallography of Membrane and Soluble Protein Micro-crystals using Synchrotron Radiation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, ... | | Authors: | Martin-Garcia, J.M, Conrad, C.E, Nelson, G, Stander, N, Zatsepin, N.A, Zook, J, Zhu, L, Geiger, J, Chun, E, Kissick, D, Hilgart, M.C, Ogata, C, Ishchenko, A, Nagaratnam, N, Roy-Chowdhury, S, Coe, J, Subramanian, G, Schaffer, A, James, D, Ketawala, G, Venugopalan, N, Xu, S, Corcoran, S, Ferguson, D, Weierstall, U, Spence, J.C.H, Cherezov, V, Fromme, P, Fischetti, R.F, Liu, W. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation.
IUCrJ, 4, 2017
|
|
6RZ7
 
 | | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2570366 (F222 space group) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{S})-8-[[4-[4-(2-chloranyl-5-fluoranyl-phenyl)butoxy]phenyl]carbonylamino]-4-(4-oxidanyl-4-oxidanylidene-butyl)-2,3- dihydro-1,4-benzoxazine-2-carboxylic acid, CHOLESTEROL, ... | | Authors: | Gusach, A, Luginina, A, Marin, E, Brouillette, R.L, Besserer-Offroy, E, Longpre, J.M, Ishchenko, A, Popov, P, Fujimoto, T, Maruyama, T, Stauch, B, Ergasheva, M, Romanovskaya, D, Stepko, A, Kovalev, K, Shevtsov, M, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors.
Nat Commun, 10, 2019
|
|
1NJK
 
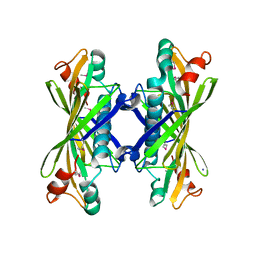 | | Crystal Structure of YbaW Probable Thioesterase from Escherichia coli | | Descriptor: | Hypothetical protein ybaW, IODIDE ION | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Xu, X, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-31 | | Release date: | 2003-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Escherichia coli Hypothetical Protein YbaW
To be Published
|
|
5UVK
 
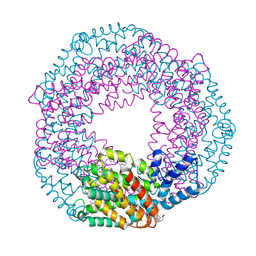 | | Serial Millisecond Crystallography of Membrane and Soluble Protein Micro-crystals using Synchrotron Radiation | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Martin-Garcia, J.M, Conrad, C.E, Nelson, G, Stander, N, Zatsepin, N.A, Zook, J, Zhu, L, Geiger, J, Chun, E, Kissick, D, Hilgart, M.C, Ogata, C, Ishchenko, A, Nagaratnam, N, Roy-Chowdhury, S, Coe, J, Subramanian, G, Schaffer, A, James, D, Ketawala, G, Venugopalan, N, Xu, S, Corcoran, S, Ferguson, D, Weierstall, U, Spence, J.C.H, Cherezov, V, Fromme, P, Fischetti, R.F, Liu, W. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-24 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation.
Iucrj, 4, 2017
|
|
5UVL
 
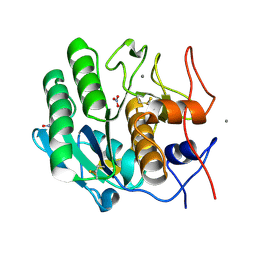 | | Serial Millisecond Crystallography of Membrane and Soluble Protein Micro-crystals using Synchrotron Radiation | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Martin-Garcia, J.M, Conrad, C.E, Nelson, G, Stander, N, Zatsepin, N.A, Zook, J, Zhu, L, Geiger, J, Chun, E, Kissick, D, Hilgart, M.C, Ogata, C, Ishchenko, A, Nagaratnam, N, Roy-Chowdhury, S, Coe, J, Subramanian, G, Schaffer, A, James, D, Ketawala, G, Venugopalan, N, Xu, S, Corcoran, S, Ferguson, D, Weierstall, U, Spence, J.C.H, Cherezov, V, Fromme, P, Fischetti, R.F, Liu, W. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation.
IUCrJ, 4, 2017
|
|
6XS4
 
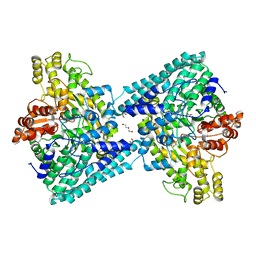 | | Crystal structure of glycyl radical enzyme ECL_02896 from Enterobacter cloacae subsp. cloacae | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Formate C-acetyltransferase | | Authors: | Valleau, D, Evdokimova, E, Stogios, P.J, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of glycyl radical enzyme ECL_02896 from Enterobacter cloacae subsp. cloacae.
To Be Published
|
|
4XVH
 
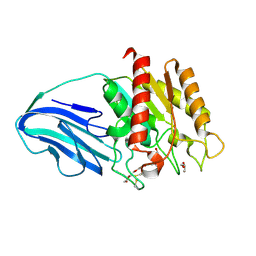 | | Crystal structure of a Corynascus thermopiles (Myceliophthora fergusii) carbohydrate esterase family 2 (CE2) enzyme plus carbohydrate binding domain (CBD) | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, Carbohydrate esterase family 2 (CE2), GLYCEROL | | Authors: | Stogios, P.J, Dong, A, Xu, X, Cui, H, Savchenko, A. | | Deposit date: | 2015-01-27 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9449 Å) | | Cite: | Crystal structure of a Corynascus thermopiles carbohydrate esterase family 2 (CE2) enzyme plus carbohydrate binding domain (CBD)
To Be Published
|
|
7SNU
 
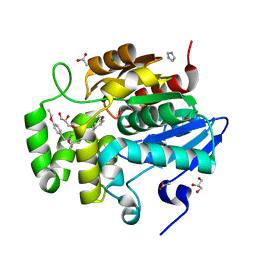 | | Crystal structure of ShHTL7 from Striga hermonthica in complex with strigolactone antagonist RG6 | | Descriptor: | 2-{(2S)-1-[(4-ethoxyphenyl)methyl]-4-[(2E)-3-(4-methoxyphenyl)prop-2-en-1-yl]piperazin-2-yl}ethan-1-ol, ACETATE ION, GLYCEROL, ... | | Authors: | Arellano-Saab, A, Stogios, P.J, Skarina, T, Yim, V, Savchenko, A, McCourt, P. | | Deposit date: | 2021-10-28 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A novel strigolactone receptor antagonist provides insights into the structural inhibition, conditioning, and germination of the crop parasite Striga.
J.Biol.Chem., 298, 2022
|
|
3E4F
 
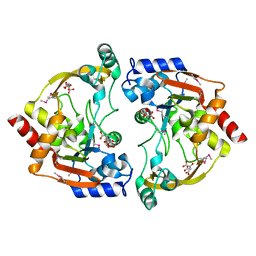 | | Crystal structure of BA2930- a putative aminoglycoside N3-acetyltransferase from Bacillus anthracis | | Descriptor: | Aminoglycoside N3-acetyltransferase, CITRIC ACID | | Authors: | Klimecka, M.M, Chruszcz, M, Skarina, T, Onopryienko, O, Cymborowski, M, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-08-11 | | Release date: | 2008-08-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
4XTK
 
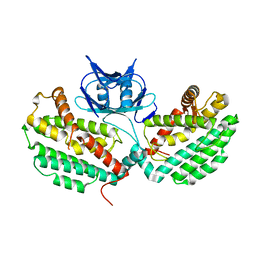 | | Structure of TM1797, a CAS1 protein from Thermotoga maritima | | Descriptor: | CRISPR-associated endonuclease Cas1 | | Authors: | Petit, P, Beloglazova, N, Skarina, T, Chang, C, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and nuclease activity of tm1797, a cas1 protein from thermotoga maritima
To Be Published
|
|
1NC7
 
 | | Crystal Structure of Thermotoga maritima 1070 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-04 | | Release date: | 2003-07-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure Analysis of Thermotoga maritima Hypothetical protein TM1070
To be Published
|
|
4XUY
 
 | | Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger | | Descriptor: | GLYCEROL, Probable endo-1,4-beta-xylanase C, SULFATE ION | | Authors: | Stogios, P.J, Dong, A, Xu, X, Cui, H, Savchenko, A. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.0027 Å) | | Cite: | Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger
To Be Published
|
|
1NI9
 
 | | 2.0 A structure of glycerol metabolism protein from E. coli | | Descriptor: | Protein glpX, SULFATE ION | | Authors: | Sanishvili, R, Brunzelle, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-23 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
