6JJS
 
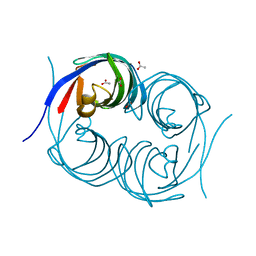 | | Crystal structure of an enzyme from Penicillium herquei in condition2 | | Descriptor: | ACETATE ION, PhnH | | Authors: | Fen, Y, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-02-26 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure and proposed mechanism of an enantioselective hydroalkoxylation enzyme from Penicillium herquei.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
6JJT
 
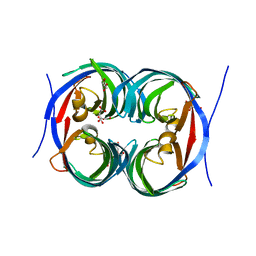 | | Crystal structure of an enzyme from Penicillium herquei in condition1 | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, PhnH | | Authors: | Fen, Y, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-02-26 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Crystal structure and proposed mechanism of an enantioselective hydroalkoxylation enzyme from Penicillium herquei.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
3J3V
 
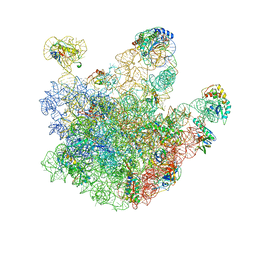 | | Atomic model of the immature 50S subunit from Bacillus subtilis (state I-a) | | Descriptor: | 50S ribosomal protein L1, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Li, N, Guo, Q, Zhang, Y, Yuan, Y, Ma, C, Lei, J, Gao, N. | | Deposit date: | 2013-04-28 | | Release date: | 2013-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (13.3 Å) | | Cite: | Cryo-EM structures of the late-stage assembly intermediates of the bacterial 50S ribosomal subunit
Nucleic Acids Res., 41, 2013
|
|
8KEC
 
 | | Cyanophage A-1(L) tail fiber | | Descriptor: | Long tail fiber, short tail fiber | | Authors: | Yu, R.C, Li, Q, Zhou, C.Z. | | Deposit date: | 2023-08-11 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the intact tail machine of Anabaena myophage A-1(L).
Nat Commun, 15, 2024
|
|
6K9C
 
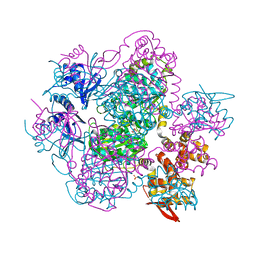 | | The apo structure of NrS-1 C terminal region (305-718) | | Descriptor: | MERCURY (II) ION, Primase, SULFATE ION | | Authors: | Chen, X, Gan, J. | | Deposit date: | 2019-06-14 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Structural studies reveal a ring-shaped architecture of deep-sea vent phage NrS-1 polymerase.
Nucleic Acids Res., 48, 2020
|
|
6JYJ
 
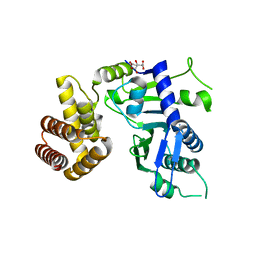 | | Crystal structure of FAM46B (TENT5B) | | Descriptor: | CITRATE ANION, Terminal nucleotidyltransferase 5B | | Authors: | Zhang, H, Hu, J.L, Gao, S. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.693199 Å) | | Cite: | FAM46B is a prokaryotic-like cytoplasmic poly(A) polymerase essential in human embryonic stem cells.
Nucleic Acids Res., 48, 2020
|
|
4PRJ
 
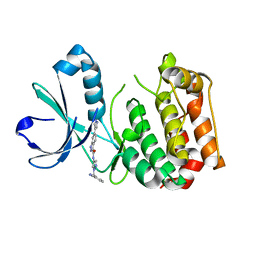 | |
5D8W
 
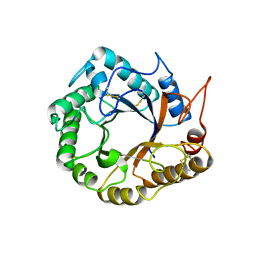 | | Structrue of a lucidum protein | | Descriptor: | Endoglucanase | | Authors: | Guo, R, Li, Q, Shang, N, Liu, G, Ko, T.P, Chen, C.C, Liu, W. | | Deposit date: | 2015-08-18 | | Release date: | 2016-06-29 | | Last modified: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Functional and structural analyses of a 1,4-beta-endoglucanase from Ganoderma lucidum.
Enzyme.Microb.Technol., 86, 2016
|
|
4ZS2
 
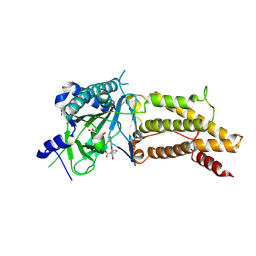 | | Structural complex of FTO/fluorescein | | Descriptor: | 2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)-BENZOIC ACID, 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase FTO, ... | | Authors: | Wang, T, Hong, T, Huang, Y, Yang, C.G, Zhou, X. | | Deposit date: | 2015-05-13 | | Release date: | 2015-11-04 | | Last modified: | 2015-11-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Fluorescein Derivatives as Bifunctional Molecules for the Simultaneous Inhibiting and Labeling of FTO Protein
J.Am.Chem.Soc., 137, 2015
|
|
4PQN
 
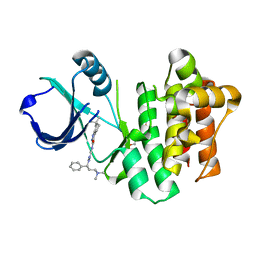 | | ITK kinase domain with compound GNE-9822 | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, N-{1-[(1S)-3-(dimethylamino)-1-phenylpropyl]-1H-pyrazol-4-yl}-6,6-dimethyl-4,5,6,7-tetrahydro-1H-indazole-3-carboxamide, Tyrosine-protein kinase ITK/TSK | | Authors: | McEwan, P.A, Barker, J.J, Eigenbrot, C. | | Deposit date: | 2014-03-03 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Property- and structure-guided discovery of a tetrahydroindazole series of interleukin-2 inducible T-cell kinase inhibitors.
J.Med.Chem., 57, 2014
|
|
4GOJ
 
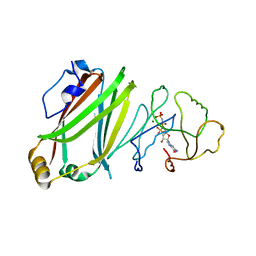 | | The Crystal Structure of full length Arl3GppNHp in complex with UNC119a | | Descriptor: | ADP-ribosylation factor-like protein 3, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Ismail, S, Xiang-Chen, Y, Miertzschke, M, Vetter, I, Koerner, C, Wittinghofer, A. | | Deposit date: | 2012-08-20 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for Arl3-specific release of myristoylated ciliary cargo from UNC119.
Embo J., 31, 2012
|
|
4GOK
 
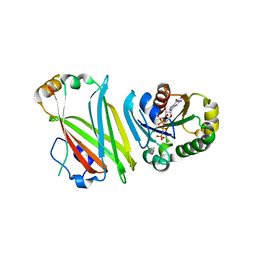 | | The Crystal structure of Arl2GppNHp in complex with UNC119a | | Descriptor: | ADP-ribosylation factor-like protein 2, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Ismail, S, Xiang-Chen, Y, Miertzschke, M, Vetter, I, Koerner, C, Wittinghofer, A. | | Deposit date: | 2012-08-20 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for Arl3-specific release of myristoylated ciliary cargo from UNC119.
Embo J., 31, 2012
|
|
1QZ4
 
 | |
1MVW
 
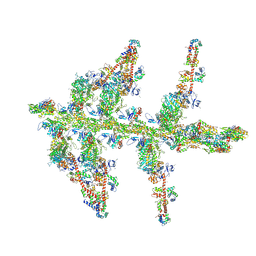 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | RABBIT SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL; LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-09-26 | | Release date: | 2002-11-20 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular Modeling of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle
J.STRUCT.BIOL., 138, 2002
|
|
6KV5
 
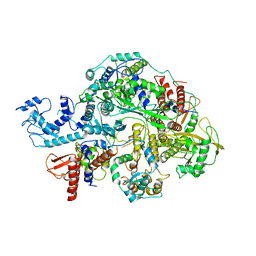 | | Structure of influenza D virus apo polymerase | | Descriptor: | Polymerase 3, Polymerase PB2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
5ZYQ
 
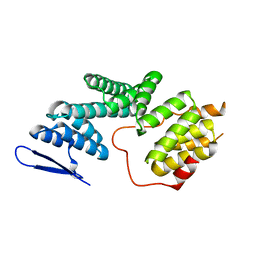 | | The Structure of Human PAF1/CTR9 complex | | Descriptor: | RNA polymerase-associated protein CTR9 homolog,RNA polymerase II-associated factor 1 homolog | | Authors: | Xie, Y, Zheng, M, Zhou, H, Long, J. | | Deposit date: | 2018-05-28 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.531 Å) | | Cite: | Paf1 and Ctr9 subcomplex formation is essential for Paf1 complex assembly and functional regulation.
Nat Commun, 9, 2018
|
|
6L4R
 
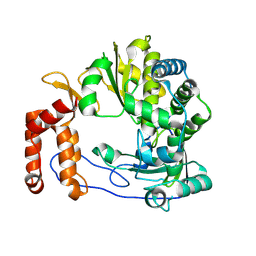 | | Crystal structure of Enterovirus D68 RdRp | | Descriptor: | RdRp | | Authors: | Wang, M.L, Li, L, Zhang, Y, Chen, Y.P, Su, D. | | Deposit date: | 2019-10-21 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Structure of the enterovirus D68 RNA-dependent RNA polymerase in complex with NADPH implicates an inhibitor binding site in the RNA template tunnel.
J.Struct.Biol., 211, 2020
|
|
1O1E
 
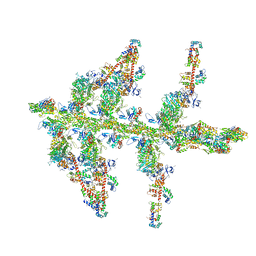 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-11-19 | | Release date: | 2002-12-04 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular Modeling of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle
J.Struct.Biol., 138, 2002
|
|
5ZYP
 
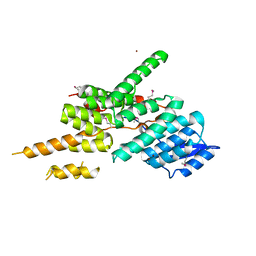 | | Structure of the Yeast Ctr9/Paf1 complex | | Descriptor: | NICKEL (II) ION, RNA polymerase-associated protein CTR9,RNA polymerase II-associated protein 1 | | Authors: | Xie, Y, Zheng, M, Zhou, H, Long, J. | | Deposit date: | 2018-05-28 | | Release date: | 2018-09-26 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (2.532 Å) | | Cite: | Paf1 and Ctr9 subcomplex formation is essential for Paf1 complex assembly and functional regulation.
Nat Commun, 9, 2018
|
|
5BNQ
 
 | | Crystal structure of hRANKL-mRANK complex | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Ren, J. | | Deposit date: | 2015-05-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A RANKL mutant used as an inter-species vaccine for efficient immunotherapy of osteoporosis.
Sci Rep, 5, 2015
|
|
7WCW
 
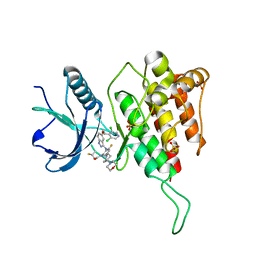 | | Crystal structure of FGFR4(V550L) kinase domain with 7v | | Descriptor: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[2-[[5-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1~{H}-indazol-3-yl]amino]-3-fluoranyl-5-(4-morpholin-4-ylpiperidin-1-yl)phenyl]propanamide | | Authors: | Chen, X.J, Lin, Q.M, Dai, S.Y, Chen, Y.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.317 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Aminoindazole Derivatives as Highly Selective Covalent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR4.
J.Med.Chem., 65, 2022
|
|
7WCX
 
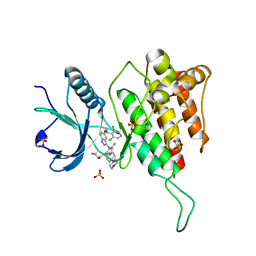 | | Crystal structure of FGFR4(V550M) kinase domain with 7v | | Descriptor: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[2-[[5-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1~{H}-indazol-3-yl]amino]-3-fluoranyl-5-(4-morpholin-4-ylpiperidin-1-yl)phenyl]propanamide | | Authors: | Chen, X.J, Lin, Q.M, Dai, S.Y, Chen, Y.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.175 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Aminoindazole Derivatives as Highly Selective Covalent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR4.
J.Med.Chem., 65, 2022
|
|
7WCT
 
 | | Crystal structure of FGFR4 kinase domain with 7v | | Descriptor: | Fibroblast growth factor receptor 4, GLYCEROL, SULFATE ION, ... | | Authors: | Chen, X.J, Lin, Q.M, Dai, S.Y, Chen, Y.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Aminoindazole Derivatives as Highly Selective Covalent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR4.
J.Med.Chem., 65, 2022
|
|
2LWF
 
 | | Structure of N-terminal domain of a plant Grx | | Descriptor: | Monothiol glutaredoxin-S16, chloroplastic | | Authors: | Feng, Y. | | Deposit date: | 2012-07-28 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the N-terminal GIY-YIG endonuclease activity of Arabidopsis glutaredoxin AtGRXS16 in chloroplasts.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4X2A
 
 | | Crystal structure of mouse glyoxalase I complexed with baicalein | | Descriptor: | 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhang, H, Zhai, J, Zhang, L, Li, C, Zhao, Y, Hu, X. | | Deposit date: | 2014-11-26 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In Vitro Inhibition of Glyoxalase І by Flavonoids: New Insights from Crystallographic Analysis.
Curr Top Med Chem, 16, 2016
|
|
