6K73
 
 | |
7EIU
 
 | | Crystal structure of Mei2 RRM3 in complex with 8mer meiRNA | | Descriptor: | GLYCEROL, Meiosis protein mei2, RNA (5'-R(P*UP*UP*CP*UP*GP*C)-3') | | Authors: | Shen, S.Y, Lv, M.Q. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Structural insights reveal the specific recognition of meiRNA by the Mei2 protein.
J Mol Cell Biol, 14, 2022
|
|
7EIO
 
 | | Crystal Structure of Mei2 RRM3 | | Descriptor: | GLYCEROL, Meiosis protein mei2, SULFATE ION | | Authors: | Shen, S.Y, Li, F.D. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Structural insights reveal the specific recognition of meiRNA by the Mei2 protein.
J Mol Cell Biol, 14, 2022
|
|
7EMJ
 
 | | Crystal structure of T2R-TTL-Barbigerone complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 8,8-dimethyl-3-(2,4,5-trimethoxyphenyl)pyrano[2,3-f]chromen-4-one, CALCIUM ION, ... | | Authors: | Yang, J.H, Yan, W. | | Deposit date: | 2021-04-14 | | Release date: | 2022-04-20 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of tubulin-barbigerone complex enables rational design of potent anticancer agents with isoflavone skeleton.
Phytomedicine, 109, 2023
|
|
6KDY
 
 | |
7EWQ
 
 | | Structure of Mumps virus nucleocapsid ring | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Su, X, Shen, Q, Shan, H. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-23 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural plasticity of mumps virus nucleocapsids with cryo-EM structures.
Commun Biol, 4, 2021
|
|
7F9I
 
 | | The apo-form structure of EnrR | | Descriptor: | EnrR repressor | | Authors: | Gan, J.H, Wang, Q.Y. | | Deposit date: | 2021-07-04 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Xenogeneic nucleoid-associated EnrR thwarts H-NS silencing of bacterial virulence with unique DNA binding.
Nucleic Acids Res., 50, 2022
|
|
7F9H
 
 | | complex structure of EnrR-DNA | | Descriptor: | EnrR repressor, target DNA | | Authors: | Gan, J.H, Wang, Q.Y. | | Deposit date: | 2021-07-04 | | Release date: | 2022-05-11 | | Last modified: | 2022-05-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Xenogeneic nucleoid-associated EnrR thwarts H-NS silencing of bacterial virulence with unique DNA binding.
Nucleic Acids Res., 50, 2022
|
|
7EUO
 
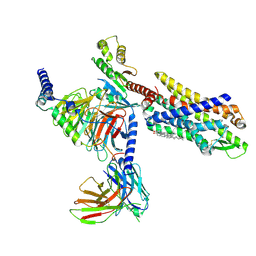 | | The structure of formyl peptide receptor 1 in complex with Gi and peptide agonist fMLF | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, X.K, Chen, G, Liao, Q.W, Du, Y, Hu, H.L, Ye, D.Q. | | Deposit date: | 2021-05-18 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for recognition of N-formyl peptides as pathogen-associated molecular patterns.
Nat Commun, 13, 2022
|
|
1QFW
 
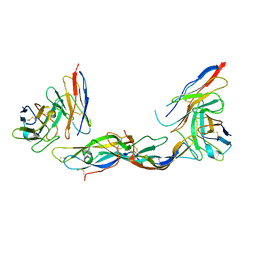 | | TERNARY COMPLEX OF HUMAN CHORIONIC GONADOTROPIN WITH FV ANTI ALPHA SUBUNIT AND FV ANTI BETA SUBUNIT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY (ANTI ALPHA SUBUNIT) (HEAVY CHAIN), ANTIBODY (ANTI ALPHA SUBUNIT) (LIGHT CHAIN), ... | | Authors: | Tegoni, M, Spinelli, S, Cambillau, C. | | Deposit date: | 1999-04-15 | | Release date: | 2000-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of a ternary complex between human chorionic gonadotropin (hCG) and two Fv fragments specific for the alpha and beta-subunits.
J.Mol.Biol., 289, 1999
|
|
7YFK
 
 | | The structure of human pregnane X receptor in complex with an SRC-1 coactivator peptide and a limonoid compound, nomilin | | Descriptor: | Nomilin, Nuclear receptor subfamily 1 group I member 2,Nuclear receptor coactivator 1 | | Authors: | Xia, Y, Yao, D, Huang, C, Cao, Y. | | Deposit date: | 2022-07-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pregnane X receptor agonist nomilin extends lifespan and healthspan in preclinical models through detoxification functions.
Nat Commun, 14, 2023
|
|
7VF6
 
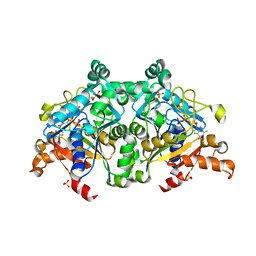 | | The crystal structure of PurZ0 | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Tong, Y, Zhang, Y. | | Deposit date: | 2021-09-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Alternative Z-genome biosynthesis pathway shows evolutionary progression from Archaea to phage.
Nat Microbiol, 8, 2023
|
|
7W0S
 
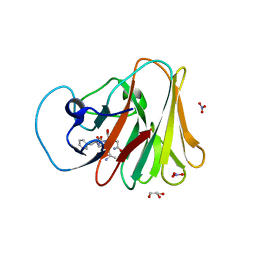 | | TRIM7 in complex with C-terminal peptide of 2C | | Descriptor: | DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase TRIM7, GLYCEROL, ... | | Authors: | Zhang, H, Liang, X, Li, X.Z. | | Deposit date: | 2021-11-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A C-terminal glutamine recognition mechanism revealed by E3 ligase TRIM7 structures.
Nat.Chem.Biol., 18, 2022
|
|
7W0Q
 
 | | TRIM7 in complex with C-terminal peptide of 2C | | Descriptor: | E3 ubiquitin-protein ligase TRIM7, peptide | | Authors: | Zhang, H, Liang, X, Li, X.Z. | | Deposit date: | 2021-11-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A C-terminal glutamine recognition mechanism revealed by E3 ligase TRIM7 structures.
Nat.Chem.Biol., 18, 2022
|
|
7W0T
 
 | | TRIM7 in complex with C-terminal peptide of 2C | | Descriptor: | E3 ubiquitin-protein ligase TRIM7, peptide | | Authors: | Zhang, H, Liang, X, Li, X.Z. | | Deposit date: | 2021-11-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A C-terminal glutamine recognition mechanism revealed by E3 ligase TRIM7 structures.
Nat.Chem.Biol., 18, 2022
|
|
7CPC
 
 | |
7CPI
 
 | |
7D4G
 
 | |
7X6Z
 
 | |
7X70
 
 | |
7X6Y
 
 | |
7DCN
 
 | | Apo-citrate lyase phosphoribosyl-dephospho-CoA transferase | | Descriptor: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, SULFATE ION, ZINC ION | | Authors: | Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
7DCM
 
 | | Crystal structure of CITX | | Descriptor: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, ZINC ION | | Authors: | Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
7DNR
 
 | |
7E5C
 
 | | Bacterial prolidase mutant D45W/L225Y/H226L/H343I | | Descriptor: | MANGANESE (II) ION, Xaa-Pro dipeptidase | | Authors: | Yang, J. | | Deposit date: | 2021-02-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-based redesign of the bacterial prolidase active-site pocket for efficient enhancement of methyl-parathion hydrolysis
Catalysis Science And Technology, 11, 2021
|
|
