5EWI
 
 | |
5F4L
 
 | | HIV-1 gp120 complex with JP-III-048 | | Descriptor: | ENVELOPE GLYCOPROTEIN GP120 of HIV-1 clade C, ~{N}'-[(1~{R},2~{R})-2-(carbamimidamidomethyl)-6-(methylaminomethyl)-2,3-dihydro-1~{H}-inden-1-yl]-~{N}-(4-chloranyl-3-fluoranyl-phenyl)ethanediamide | | Authors: | Liang, S, Hendrickson, W.A. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-30 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Small-Molecule CD4-Mimics: Structure-Based Optimization of HIV-1 Entry Inhibition.
Acs Med.Chem.Lett., 7, 2016
|
|
5F9O
 
 | | Crystal structure of broadly neutralizing VH1-46 germline-derived CD4-binding site-directed antibody CH235.09 in complex with HIV-1 clade A/E 93TH057 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CH235.09 Light chain, ... | | Authors: | Joyce, M.G, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody.
Cell, 165, 2016
|
|
5F89
 
 | |
7ZFS
 
 | | BRD4 in complex with FragLite32 | | Descriptor: | 1,2-ETHANEDIOL, 4-bromanyl-1,2-dimethoxy-benzene, Bromodomain-containing protein 4, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZFZ
 
 | | BRD4 in complex with PepLite-Val | | Descriptor: | (2R)-2-acetamido-N-(3-bromanylprop-2-ynyl)-3-methyl-butanamide, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZFV
 
 | | BRD4 in complex with PepLite-Ala | | Descriptor: | (2~{S})-2-acetamido-~{N}-(3-bromanylprop-2-ynyl)propanamide, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZFY
 
 | | BRD4 in complex with PepLite-Gly | | Descriptor: | 2-acetamido-N-(3-bromanylprop-2-ynyl)ethanamide, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZFO
 
 | | BRD4 in complex with FragLite28 | | Descriptor: | 4-bromo-1-(2-hydroxyethyl)pyridin-2(1H)-one, GLYCEROL, Isoform C of Bromodomain-containing protein 4 | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZFN
 
 | | BRD4 in complex with FragLite24 | | Descriptor: | 4-bromanyl-2-oxidanyl-benzoic acid, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZFU
 
 | | BRD4 in complex with PepLite-Pro | | Descriptor: | (2R)-N-(3-bromanylprop-2-ynyl)-1-ethanoyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZFT
 
 | | BRD4 in complex with FragLite33 | | Descriptor: | 3-azanyl-5-bromanyl-1-methyl-pyridin-2-one, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZG1
 
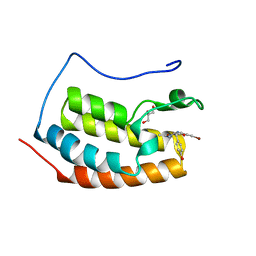 | | BRD4 in complex with PepLite-Tyr | | Descriptor: | Bromodomain-containing protein 4, GLYCEROL, Nalpha-acetyl-N-(3-bromoprop-2-yn-1-yl)-L-tyrosinamide | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZG2
 
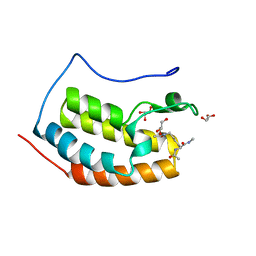 | | BRD4 in complex with Acetyl-Lys | | Descriptor: | (2S)-2,6-diacetamido-N-methyl-hexanamide, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
5ESZ
 
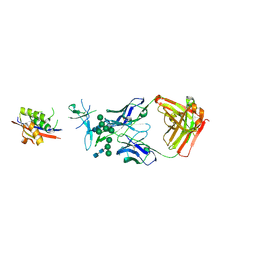 | | Crystal Structure of Broadly Neutralizing Antibody CH04, Isolated from Donor CH0219, in Complex with Scaffolded Trimeric HIV-1 Env V1V2 Domain from the Clade AE Strain A244 | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase,Envelope glycoprotein gp160, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH04 Heavy Chain, ... | | Authors: | Gorman, J, Yang, M, Kwong, P.D. | | Deposit date: | 2015-11-17 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.191 Å) | | Cite: | Structures of HIV-1 Env V1V2 with broadly neutralizing antibodies reveal commonalities that enable vaccine design.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5ESV
 
 | | Crystal Structure of Broadly Neutralizing Antibody CH03, Isolated from Donor CH0219, in Complex with Scaffolded Trimeric HIV-1 Env V1V2 Domain from the Clade C Superinfecting Strain of Donor CAP256. | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase,Envelope glycoprotein gp160, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorman, J, Yang, M, Kwong, P.D. | | Deposit date: | 2015-11-17 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.105 Å) | | Cite: | Structures of HIV-1 Env V1V2 with broadly neutralizing antibodies reveal commonalities that enable vaccine design.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5F96
 
 | |
5FBN
 
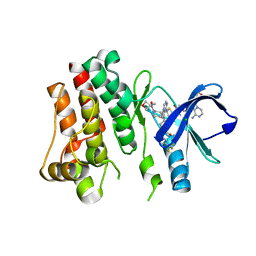 | | BTK kinase domain with inhibitor 1 | | Descriptor: | 1,2-ETHANEDIOL, 4-[8-azanyl-3-[(2~{S})-1-[4-(dimethylamino)butanoyl]pyrrolidin-2-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-(1,3-thiazol-2-yl)benzamide, 4-[8-azanyl-3-[(3~{R})-1-(3-methyloxetan-3-yl)carbonylpiperidin-3-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-[4-(trifluoromethyl)pyridin-2-yl]benzamide, ... | | Authors: | Raaijmakers, H.C.A, Vu-Pham, D. | | Deposit date: | 2015-12-14 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of 8-Amino-imidazo[1,5-a]pyrazines as Reversible BTK Inhibitors for the Treatment of Rheumatoid Arthritis.
Acs Med.Chem.Lett., 7, 2016
|
|
5EOL
 
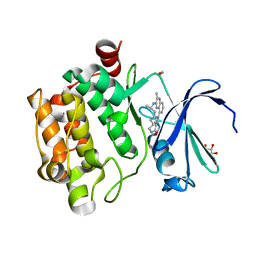 | |
7Z3A
 
 | | AMC009 SOSIPv5.2 in complex with Fabs ACS101 and ACS124 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACS101 heavy chain, ... | | Authors: | van Schooten, J, Ward, A. | | Deposit date: | 2022-03-02 | | Release date: | 2022-08-10 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
5EIV
 
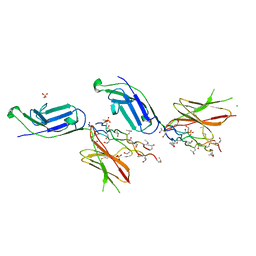 | | Crystal structure of complex of osteoclast-associated immunoglobulin-like receptor (OSCAR) and a synthetic collagen consensus peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CHLORIDE ION, ... | | Authors: | Zhou, L, Blaszczyk, M, Chirgadze, D, Bihan, D, Farndale, R.W. | | Deposit date: | 2015-10-30 | | Release date: | 2015-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.414 Å) | | Cite: | Structural basis for collagen recognition by the immune receptor OSCAR.
Blood, 127, 2016
|
|
7ZHN
 
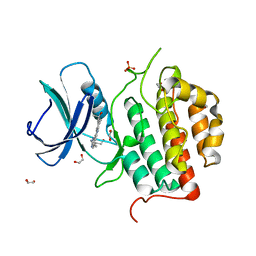 | | Crystal structure of TTBK1 in complex with AMG28 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-amino-5,6,7,8-tetrahydropyrimido[4',5':3,4]cyclohepta[1,2-b]indol-11-yl)-2-methylbut-3-yn-2-ol, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Axtman, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Modulation of tau tubulin kinases (TTBK1 and TTBK2) impacts ciliogenesis.
Sci Rep, 13, 2023
|
|
7ZHQ
 
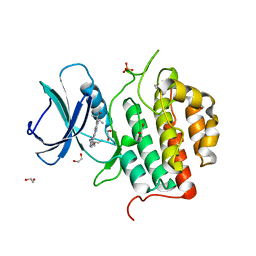 | | Crystal structure of TTBK1 in complex with compound 10 (7-009) | | Descriptor: | (3~{S})-1-(4-azanyl-3,5,12-triazatetracyclo[9.7.0.0^{2,7}.0^{13,18}]octadeca-1(11),2,4,6,13(18),14,16-heptaen-16-yl)-3-methyl-pent-1-yn-3-ol, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Axtman, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of tau tubulin kinases (TTBK1 and TTBK2) impacts ciliogenesis.
Sci Rep, 13, 2023
|
|
7ZHO
 
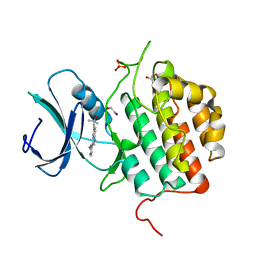 | | Crystal structure of TTBK1 in complex with compound 3 (7-001) | | Descriptor: | 1,2-ETHANEDIOL, 4-[3-(2-azanylpyrimidin-4-yl)-1~{H}-indol-5-yl]-2-methyl-but-3-yn-2-ol, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Axtman, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Modulation of tau tubulin kinases (TTBK1 and TTBK2) impacts ciliogenesis.
Sci Rep, 13, 2023
|
|
7ZHP
 
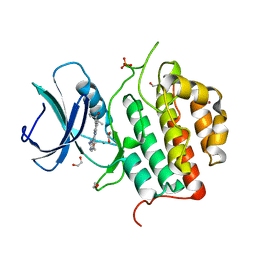 | | Crystal structure of TTBK1 in complex with compound 9 (7-005) | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-azanyl-3,5,12-triazatetracyclo[9.7.0.0^{2,7}.0^{13,18}]octadeca-1(11),2,4,6,13(18),14,16-heptaen-16-yl)-3-ethyl-pent-1-yn-3-ol, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Axtman, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of tau tubulin kinases (TTBK1 and TTBK2) impacts ciliogenesis.
Sci Rep, 13, 2023
|
|
