5OX5
 
 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with CCT6, a GSK1278863-related compound | | Descriptor: | (6-hydroxy-1,3-dimethyl-2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carbonyl)glycine, BICARBONATE ION, Egl nine homolog 1, ... | | Authors: | Chowdhury, R, Thinnes, C.C, Schofield, C.J. | | Deposit date: | 2017-09-06 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Molecular and cellular mechanisms of HIF prolyl hydroxylase inhibitors in clinical trials.
Chem Sci, 8, 2017
|
|
4WWN
 
 | | Crystal structure of human PI3K-gamma in complex with (S)-N-(1-(7-fluoro-2-(pyridin-2-yl)quinolin-3-yl)ethyl)-9H-purin-6-amine AMG319 inhibitor | | Descriptor: | N-{(1S)-1-[7-fluoro-2-(pyridin-2-yl)quinolin-3-yl]ethyl}-9H-purin-6-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2014-11-11 | | Release date: | 2014-12-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and in Vivo Evaluation of (S)-N-(1-(7-Fluoro-2-(pyridin-2-yl)quinolin-3-yl)ethyl)-9H-purin-6-amine (AMG319) and Related PI3K delta Inhibitors for Inflammation and Autoimmune Disease.
J.Med.Chem., 58, 2015
|
|
4WWO
 
 | | Crystal structure of human PI3K-gamma in complex with phenylquinoline inhibitor N-{(1S)-1-[8-chloro-2-(3-fluorophenyl)quinolin-3-yl]ethyl}-9H-purin-6-amine | | Descriptor: | N-{(1S)-1-[8-chloro-2-(3-fluorophenyl)quinolin-3-yl]ethyl}-9H-purin-6-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2014-11-11 | | Release date: | 2014-12-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and in Vivo Evaluation of (S)-N-(1-(7-Fluoro-2-(pyridin-2-yl)quinolin-3-yl)ethyl)-9H-purin-6-amine (AMG319) and Related PI3K delta Inhibitors for Inflammation and Autoimmune Disease.
J.Med.Chem., 58, 2015
|
|
3KU0
 
 | | Structure of GAP31 with adenine at its binding pocket | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Ribosome-inactivating protein gelonin | | Authors: | Kong, X.-P. | | Deposit date: | 2009-11-26 | | Release date: | 2010-01-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new activity of anti-HIV and anti-tumor protein GAP31: DNA adenosine glycosidase--structural and modeling insight into its functions.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
6LIS
 
 | | ASFV dUTPase in complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, E165R | | Authors: | Liang, R, Peng, G.Q. | | Deposit date: | 2019-12-12 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development.
J.Biol.Chem., 296, 2020
|
|
6LJ3
 
 | |
6LJO
 
 | | African swine fever virus dUTPase | | Descriptor: | E165R | | Authors: | Liang, R, Peng, G.Q. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development.
J.Biol.Chem., 296, 2020
|
|
6MC4
 
 | |
6M79
 
 | |
7XF3
 
 | | The structure of HLA-B*1501/BM58-66AF9 | | Descriptor: | 9-mer peptide from Matrix protein 1, Beta-2-microglobulin, MHC class I antigen | | Authors: | Zhao, Y.Z, Xiao, W.L, Wu, Y.N, Fan, W.F, Yue, C, Zhang, Q.X, Zhang, D.N, Yuan, X.J, Yao, S.J, Liu, S, Li, M, Wang, P.Y, Zhang, H.J, Zhang, J, Zhao, M, Zheng, X.Q, Liu, W.J, Gao, G.F, Liu, W.L. | | Deposit date: | 2022-03-31 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Parallel T Cell Immunogenic Regions in Influenza B and A Viruses with Distinct Nuclear Export Signal Functions: The Balance between Viral Life Cycle and Immune Escape.
J Immunol., 210, 2023
|
|
5XKX
 
 | | Crystal structure of WT DddY | | Descriptor: | ACETATE ION, Uncharacterized protein, ZINC ION | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2017-05-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Insights into Dimethylsulfoniopropionate Lyase DddY, a New Member of the Cupin Superfamily.
J. Mol. Biol., 429, 2017
|
|
5Y4K
 
 | | Crystal structure of DddY mutant Y260A | | Descriptor: | ACRYLIC ACID, DMSP lyase DddY, ZINC ION | | Authors: | Zhang, Y.Z, Li, C.Y. | | Deposit date: | 2017-08-03 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic Insights into Dimethylsulfoniopropionate Lyase DddY, a New Member of the Cupin Superfamily.
J. Mol. Biol., 429, 2017
|
|
5XKY
 
 | | Crystal structure of DddY Se derivative | | Descriptor: | Uncharacterized protein, ZINC ION | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2017-05-10 | | Release date: | 2017-11-01 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Mechanistic Insights into Dimethylsulfoniopropionate Lyase DddY, a New Member of the Cupin Superfamily.
J. Mol. Biol., 429, 2017
|
|
9BJU
 
 | | Crystal structure of the complex between VHL, ElonginB, ElonginC, and compound 5 | | Descriptor: | 1,2-ETHANEDIOL, 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, ACETATE ION, ... | | Authors: | Murray, J.M, Wu, H, Fuhrmann, J, Fairbrother, W.J, DiPasquale, A. | | Deposit date: | 2024-04-25 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Potency-Enhanced Peptidomimetic VHL Ligands with Improved Oral Bioavailability.
J.Med.Chem., 67, 2024
|
|
9BOL
 
 | | Crystal structure of the complex between VHL, ElonginB, ElonginC, and compound 5 | | Descriptor: | (4R)-1-[(2S)-2-(4-cyclopropyl-1H-1,2,3-triazol-1-yl)-3,3-dimethylbutanoyl]-4-hydroxy-N-{[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl}-L-prolinamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Murray, J.M, Wu, H, Fuhrmann, J, Fairbrother, W.J, DiPasquale, A. | | Deposit date: | 2024-05-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Potency-Enhanced Peptidomimetic VHL Ligands with Improved Oral Bioavailability.
J.Med.Chem., 67, 2024
|
|
8Z9Z
 
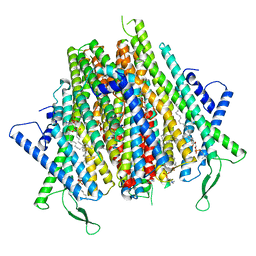 | | Cryo-EM structure of the insect olfactory receptor OR5-Orco heterocomplex from Acyrthosiphon pisum | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Odorant receptor, ApisOR5, ... | | Authors: | Wang, Y.D, Qiu, L, Guan, Z.Y, Wang, Q, Wang, G.R, Yin, P. | | Deposit date: | 2024-04-24 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for odorant recognition of the insect odorant receptor OR-Orco heterocomplex.
Science, 384, 2024
|
|
8Z9A
 
 | | Cryo-EM structure of the insect olfactory receptor OR5-Orco heterocomplex from Acyrthosiphon pisum bound with geranyl acetate | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Odorant receptor, ApisOR5, ... | | Authors: | Wang, Y.D, Qiu, L, Guan, Z.Y, Wang, Q, Wang, G.R, Yin, P. | | Deposit date: | 2024-04-23 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for odorant recognition of the insect odorant receptor OR-Orco heterocomplex.
Science, 384, 2024
|
|
6PTJ
 
 | | Structure of Ctf4 trimer in complex with one CMG helicase | | Descriptor: | Cell division control protein 45, DNA polymerase alpha-binding protein, DNA replication complex GINS protein PSF1, ... | | Authors: | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
6PTO
 
 | | Structure of Ctf4 trimer in complex with three CMG helicases | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA polymerase alpha-binding protein, ... | | Authors: | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | Deposit date: | 2019-07-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
6PTN
 
 | | Structure of Ctf4 trimer in complex with two CMG helicases | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA polymerase alpha-binding protein, ... | | Authors: | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | Deposit date: | 2019-07-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
8E7O
 
 | | CRYSTAL STRUCTURE OF LYS48-LINKED TETRAUBIQUITIN | | Descriptor: | SULFATE ION, Ubiquitin | | Authors: | Lemma, B.E, Fushman, D. | | Deposit date: | 2022-08-24 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of selective recognition of Lys48-linked polyubiquitin by macrocyclic peptide inhibitors of proteasomal degradation.
Nat Commun, 14, 2023
|
|
5YGU
 
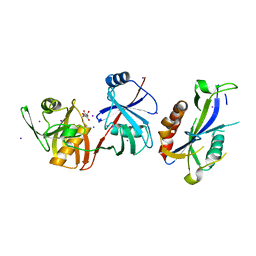 | | Crystal structure of Escherichia coli (strain K12) mRNA Decapping Complex RppH-DapF | | Descriptor: | Diaminopimelate epimerase, IODIDE ION, L(+)-TARTARIC ACID, ... | | Authors: | Wang, Q, Guan, Z.Y, Zhang, D.L, Zou, T.T, Yin, P. | | Deposit date: | 2017-09-27 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | DapF stabilizes the substrate-favoring conformation of RppH to stimulate its RNA-pyrophosphohydrolase activity in Escherichia coli.
Nucleic Acids Res., 46, 2018
|
|
6JJB
 
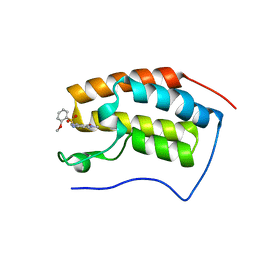 | | BRD4 in complex with ZZM1 | | Descriptor: | 2-methoxy-N-(1-methyl-2-oxidanylidene-benzo[cd]indol-6-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Xu, J, Chen, Y, Jiang, F, Zhu, J. | | Deposit date: | 2019-02-25 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.508 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones and Pyrrolo[4,3,2-de]quinolin-2(1H)-ones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with Selectivity for the First Bromodomain with Potential High Efficiency against Acute Gouty Arthritis.
J.Med.Chem., 62, 2019
|
|
5YZV
 
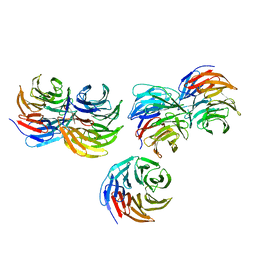 | | Biophysical and structural characterization of the thermostable WD40 domain of a prokaryotic protein, Thermomonospora curvata PkwA | | Descriptor: | Probable serine/threonine-protein kinase PkwA | | Authors: | Li, D.Y, Shen, C, Du, Y, Qiao, F.F, Kong, T, Yuan, L.R, Zhang, D.L, Wu, X.H, Wu, Y.D. | | Deposit date: | 2017-12-15 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biophysical and structural characterization of the thermostable WD40 domain of a prokaryotic protein, Thermomonospora curvata PkwA
Sci Rep, 8, 2018
|
|
6JJ3
 
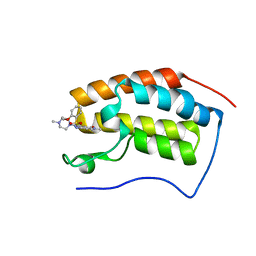 | | BRD4 in complex with 138A | | Descriptor: | 2-methoxy-N-[2-methyl-6-(4-methylpiperazin-1-yl)-3-oxidanylidene-2,7-diazatricyclo[6.3.1.0^{4,12}]dodeca-1(12),4,6,8,10-pentaen-9-yl]benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Xu, J, Chen, Y, Jiang, F, Zhu, J. | | Deposit date: | 2019-02-25 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.718 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones and Pyrrolo[4,3,2-de]quinolin-2(1H)-ones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with Selectivity for the First Bromodomain with Potential High Efficiency against Acute Gouty Arthritis.
J.Med.Chem., 62, 2019
|
|
