3KW4
 
 | | Crystal structure of cytochrome 2B4 in complex with the anti-platelet drug ticlopidine | | Descriptor: | 2-{[(3alpha,5alpha,7alpha,8alpha,10alpha,12alpha,17alpha)-3,12-bis{2-[(4-O-alpha-D-glucopyranosyl-beta-D-glucopyranosyl)oxy]ethoxy}cholan-7-yl]oxy}ethyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, ... | | Authors: | Gay, S.C, Maekawa, K, Roberts, A.G, Hong, W.-X, Zhang, Q, Stout, C.D, Halpert, J.R. | | Deposit date: | 2009-11-30 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structures of cytochrome P450 2B4 complexed with the antiplatelet drugs ticlopidine and clopidogrel.
Biochemistry, 49, 2010
|
|
5KT5
 
 | | Teranry complex of human DNA polymerase iota R96G inserting dCMPNPP opposite template G in the presence of Mn2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*CP*TP*GP*GP*GP*GP*TP*CP*CP*T)-3'), DNA (5'-D(P*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Choi, J.Y, Patra, A, Yeom, M, Lee, Y.S, Zhang, Q, Egli, M, Guengerich, F.P. | | Deposit date: | 2016-07-11 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Kinetic and Structural Impact of Metal Ions and Genetic Variations on Human DNA Polymerase iota.
J.Biol.Chem., 291, 2016
|
|
6RFL
 
 | | Structure of the complete Vaccinia DNA-dependent RNA polymerase complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Grimm, C, Hillen, S.H, Bedenk, K, Bartuli, J, Neyer, S, Zhang, Q, Huettenhofer, A, Erlacher, M, Dienemann, C, Schlosser, A, Urlaub, H, Boettcher, B, Szalay, A.A, Cramer, P, Fischer, U. | | Deposit date: | 2019-04-15 | | Release date: | 2019-12-11 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Vaccinia RNA Polymerase Complexes.
Cell, 179, 2019
|
|
5VBL
 
 | | Structure of apelin receptor in complex with agonist peptide | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apelin receptor,Rubredoxin,Apelin receptor Chimera, ZINC ION, ... | | Authors: | Ma, Y, Yue, Y, Ma, Y, Zhang, Q, Zhou, Q, Song, Y, Shen, Y, Li, X, Ma, X, Li, C, Hanson, M.A, Han, G.W, Sickmier, E.A, Swaminath, G, Zhao, S, Stevems, R.C, Hu, L.A, Zhong, W, Zhang, M, Xu, F. | | Deposit date: | 2017-03-29 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Apelin Control of the Human Apelin Receptor
Structure, 25, 2017
|
|
3QPS
 
 | | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni | | Descriptor: | CHOLIC ACID, CmeR | | Authors: | Lei, H.T, Routh, M.D, Shen, Z, Su, C.C, Zhang, Q, Yu, E.W. | | Deposit date: | 2011-02-14 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni.
Protein Sci., 20, 2011
|
|
3QQA
 
 | | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni | | Descriptor: | CmeR, TAUROCHOLIC ACID | | Authors: | Lei, H.T, Routh, M.D, Shen, Z, Su, C.-C, Zhang, Q, Yu, E.W. | | Deposit date: | 2011-02-15 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni.
Protein Sci., 20, 2011
|
|
3SYY
 
 | | Crystal Structure of an alkaline exonuclease (LHK-Exo) from Laribacter hongkongensis | | Descriptor: | Exonuclease, MAGNESIUM ION | | Authors: | Yang, W, Chen, W.Y, Wang, H, Zhang, Q, Zhou, W, Bartlam, M, Watt, R.M, Rao, Z. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insight into the mechanism of an alkaline exonuclease from Laribacter hongkongensis.
Nucleic Acids Res., 39, 2011
|
|
3SZ4
 
 | | Crystal Structure of LHK-Exo in complex with dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Exonuclease, MAGNESIUM ION | | Authors: | Yang, W, Chen, W.Y, Wang, H, Zhang, Q, Zhou, W, Bartlam, M, Watt, R.M, Rao, Z. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural and functional insight into the mechanism of an alkaline exonuclease from Laribacter hongkongensis.
Nucleic Acids Res., 39, 2011
|
|
5XKA
 
 | | Crystal structure of M.tuberculosis PknI kinase domain | | Descriptor: | Serine/threonine-protein kinase PknI | | Authors: | Yan, Q, Jiang, D, Qian, L, Zhang, Q, Zhang, W, Zhou, W, Mi, K, Guddat, L, Yang, H, Rao, Z. | | Deposit date: | 2017-05-06 | | Release date: | 2018-05-16 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structural Insight into the Activation of PknI Kinase from M. tuberculosis via Dimerization of the Extracellular Sensor Domain.
Structure, 25, 2017
|
|
7JO9
 
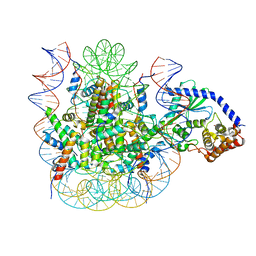 | | 1:1 cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1, ... | | Authors: | Boyer, J.A, Spangler, C.J, Strauss, J.D, Cesmat, A.P, Liu, P, McGinty, R.K, Zhang, Q. | | Deposit date: | 2020-08-06 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of nucleosome-dependent cGAS inhibition.
Science, 370, 2020
|
|
7JOA
 
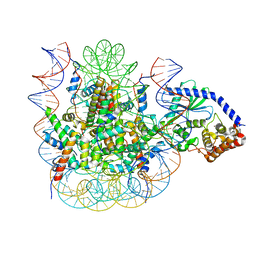 | | 2:1 cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1, ... | | Authors: | Boyer, J.A, Spangler, C.J, Strauss, J.D, Cesmat, A.P, Liu, P, McGinty, R.K, Zhang, Q. | | Deposit date: | 2020-08-06 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of nucleosome-dependent cGAS inhibition.
Science, 370, 2020
|
|
4HYX
 
 | | Crystal Structure Analysis of the Bacteriorhodopsin in Facial Amphiphile-4 DMPC Bicelle | | Descriptor: | Bacteriorhodopsin, DECANE, GLYCEROL, ... | | Authors: | Lee, S, Stout, C.D, Zhang, Q. | | Deposit date: | 2012-11-14 | | Release date: | 2013-03-20 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HWL
 
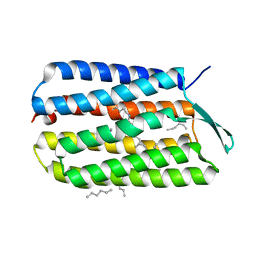 | | Crystal Structure Analysis of the Bacteriorhodopsin in Facial Amphiphile-7 DMPC Bicelle | | Descriptor: | Bacteriorhodopsin, GLYCEROL, HEPTANE, ... | | Authors: | Lee, S, Stout, C.D, Zhang, Q. | | Deposit date: | 2012-11-08 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4F4C
 
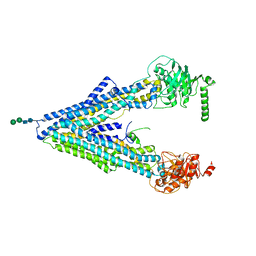 | | The Crystal Structure of the Multi-Drug Transporter | | Descriptor: | Multidrug resistance protein pgp-1, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, undecyl 4-O-alpha-D-glucopyranosyl-1-thio-beta-D-glucopyranoside | | Authors: | Jin, M.S, Oldham, M.L, Zhang, Q, Chen, J. | | Deposit date: | 2012-05-10 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans.
Nature, 490, 2012
|
|
5XPW
 
 | | Structure of amphioxus IgVJ-C2 molecule | | Descriptor: | amphioxus IgVJ-C2 | | Authors: | Chen, R, Qi, J, Zhang, N, Zhang, L, Yao, S, Wu, Y, Jiang, B, Wang, Z, Yuan, H, Zhang, Q, Xia, C. | | Deposit date: | 2017-06-05 | | Release date: | 2018-04-18 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Discovery and Analysis of Invertebrate IgVJ-C2 Structure from Amphioxus Provides Insight into the Evolution of the Ig Superfamily.
J. Immunol., 200, 2018
|
|
6BL6
 
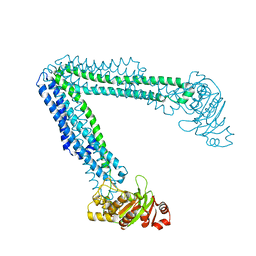 | | Crystallization of lipid A transporter MsbA from Salmonella typhimurium | | Descriptor: | Lipid A export ATP-binding/permease protein MsbA | | Authors: | Padayatti, P.S, Stanfield, R.L, Zhang, Q, Wilson, I.A, Lee, S.C. | | Deposit date: | 2017-11-09 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Lipid A Transport Pathway in MsbA.
Structure, 27, 2019
|
|
6BNH
 
 | | Solution NMR structures of BRD4 ET domain with JMJD6 peptide | | Descriptor: | Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, Bromodomain-containing protein 4 | | Authors: | Konuma, T, Yu, D, Zhao, C, Ju, Y, Sharma, R, Ren, C, Zhang, Q, Zhou, M.-M, Zeng, L. | | Deposit date: | 2017-11-16 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Mechanism of the Oxygenase JMJD6 Recognition by the Extraterminal (ET) Domain of BRD4.
Sci Rep, 7, 2017
|
|
5XPV
 
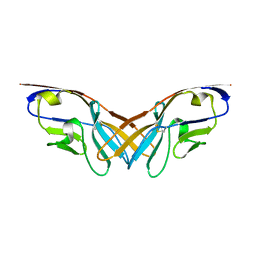 | | Structure of the V domain of amphioxus IgVJ-C2 | | Descriptor: | amphioxus IgVJ-C2 | | Authors: | Chen, R, Qi, J, Zhang, N, Zhang, L, Yao, S, Wu, Y, Jiang, B, Wang, Z, Yuan, H, Zhang, Q, Xia, C. | | Deposit date: | 2017-06-05 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Analysis of Invertebrate IgVJ-C2 Structure from Amphioxus Provides Insight into the Evolution of the Ig Superfamily.
J. Immunol., 200, 2018
|
|
6W8T
 
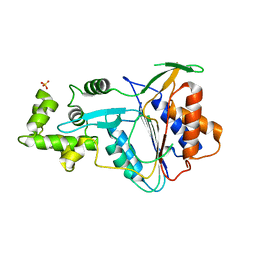 | | Crystal structure of metacaspase 4 from Arabidopsis (microcrystals treated with calcium) | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
6W8R
 
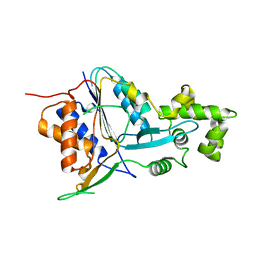 | | Crystal structure of metacaspase 4 C139A from Arabidopsis | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
6W8S
 
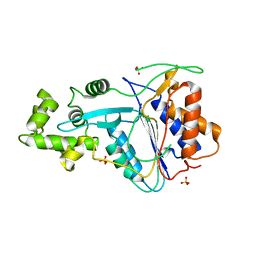 | | Crystal structure of metacaspase 4 from Arabidopsis | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.484 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
4B03
 
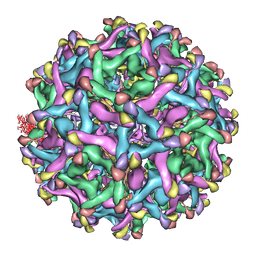 | | 6A Electron cryomicroscopy structure of immature Dengue virus serotype 1 | | Descriptor: | DENGUE VIRUS 1 E PROTEIN, DENGUE VIRUS 1 PRM PROTEIN | | Authors: | Kostyuchenko, V.A, Zhang, Q, Tan, L.C, Ng, T.S, Lok, S.M. | | Deposit date: | 2012-06-28 | | Release date: | 2013-06-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Immature and Mature Dengue Serotype 1 Virus Structures Provide Insight Into the Maturation Process.
J.Virol., 87, 2013
|
|
4CCT
 
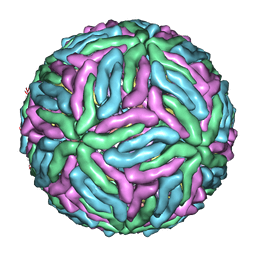 | | Dengue 1 cryo-EM reconstruction | | Descriptor: | DENGUE VIRUS 1 E PROTEIN, DENGUE VIRUS 1 M PROTEIN | | Authors: | Kostyuchenko, V.A, Zhang, Q, Tan, J.L, Ng, T.S, Lok, S.M. | | Deposit date: | 2013-10-28 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Immature and Mature Dengue Serotype 1 Virus Structures Provide Insight Into the Maturation Process.
J.Virol., 87, 2013
|
|
8W9W
 
 | | The cryo-EM structure of human sphingomyelin synthase-related protein in complex with ceramide/phosphoethanolamine | | Descriptor: | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, Sphingomyelin synthase-related protein 1, ~{N}-[(~{Z},2~{S},3~{R})-1,3-bis(oxidanyl)heptadec-4-en-2-yl]dodecanamide | | Authors: | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | Deposit date: | 2023-09-06 | | Release date: | 2024-02-28 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8W9Y
 
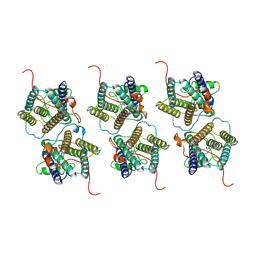 | | The cryo-EM structure of human sphingomyelin synthase-related protein | | Descriptor: | Sphingomyelin synthase-related protein 1 | | Authors: | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | Deposit date: | 2023-09-06 | | Release date: | 2024-02-28 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
