7WC5
 
 | | Crystal structure of serotonin 2A receptor in complex with psilocin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-(dimethylamino)ethyl]-1~{H}-indol-4-ol, 5-hydroxytryptamine receptor 2A, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC6
 
 | | Crystal structure of serotonin 2A receptor in complex with LSD | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC9
 
 | | Crystal structure of serotonin 2A receptor in complex with non-hallucinogenic psychedelic analog | | Descriptor: | (10~{R},15~{S})-12-[3-(2-methoxyphenyl)propyl]-4-methyl-1,4,12-triazatetracyclo[7.6.1.0^{5,16}.0^{10,15}]hexadeca-5(16),6,8-triene, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC7
 
 | | Crystal structure of serotonin 2A receptor in complex with lisuride | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, CHOLESTEROL, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC8
 
 | | Crystal structure of serotonin 2A receptor in complex with lumateperone | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(4-fluorophenyl)-4-[(10~{R},15~{S})-4-methyl-1,4,12-triazatetracyclo[7.6.1.0^{5,16}.0^{10,15}]hexadeca-5,7,9(16)-trien-12-yl]butan-1-one, 5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
8PKP
 
 | | Cryo-EM structure of the apo Anaphase-promoting complex/cyclosome (APC/C) at 3.2 Angstrom resolution | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Hoefler, A, Yu, J, Chang, L, Zhang, Z, Yang, J, Boland, A, Barford, D. | | Deposit date: | 2023-06-27 | | Release date: | 2023-07-19 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of apo-APC/C and APC/C CDH1:EMI1 complexes provide insights into APC/C regulation.
Nat Commun, 15, 2024
|
|
7VOE
 
 | | Crystal structure of 5-HT2AR in complex with aripiprazole | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, 7-[4-[4-[2,3-bis(chloranyl)phenyl]piperazin-1-yl]butoxy]-3,4-dihydro-1H-quinolin-2-one, ... | | Authors: | Chen, Z, Fan, L, Wang, H, Yu, J, Lu, D, Qi, J, Nie, F, Luo, Z, Liu, Z, Cheng, J, Wang, S. | | Deposit date: | 2021-10-13 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based design of a novel third-generation antipsychotic drug lead with potential antidepressant properties.
Nat.Neurosci., 25, 2022
|
|
7VOD
 
 | | Crystal structure of 5-HT2AR in complex with cariprazine | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[4-[2-[4-[2,3-bis(chloranyl)phenyl]piperazin-1-yl]ethyl]cyclohexyl]-1,1-dimethyl-urea, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Chen, Z, Fan, L, Wang, H, Yu, J, Lu, D, Qi, J, Nie, F, Luo, Z, Liu, Z, Cheng, J, Wang, S. | | Deposit date: | 2021-10-13 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-based design of a novel third-generation antipsychotic drug lead with potential antidepressant properties.
Nat.Neurosci., 25, 2022
|
|
2KSM
 
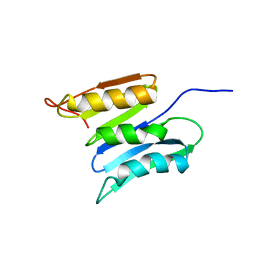 | | Central B domain of Rv0899 from Mycobacterium tuberculosis | | Descriptor: | MYCOBACTERIUM TUBERCULOSIS RV0899/MT0922/OmpATb | | Authors: | Teriete, P, Yao, Y, Kolodzik, A, Yu, J, Song, H, Niederweis, M, Marassi, F.M. | | Deposit date: | 2010-01-07 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mycobacterium tuberculosis Rv0899 adopts a mixed alpha/beta-structure and does not form a transmembrane beta-barrel.
Biochemistry, 49, 2010
|
|
4DC2
 
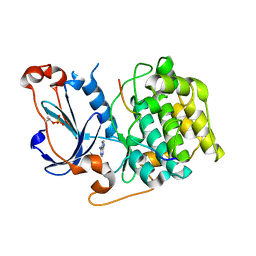 | | Structure of PKC in Complex with a Substrate Peptide from Par-3 | | Descriptor: | ADENINE, Partitioning defective 3 homolog, Protein kinase C iota type | | Authors: | Shang, Y, Wang, C, Yu, J, Zhang, M. | | Deposit date: | 2012-01-17 | | Release date: | 2012-07-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Substrate recognition mechanism of atypical protein kinase Cs revealed by the structure of PKC iota in complex with a substrate peptide from Par-3
Structure, 20, 2012
|
|
8H1L
 
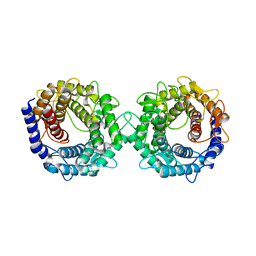 | | Crystal structure of glucose-2-epimerase in complex with D-Glucitol from Runella slithyformis Runsl_4512 | | Descriptor: | N-acylglucosamine 2-epimerase, sorbitol | | Authors: | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H1K
 
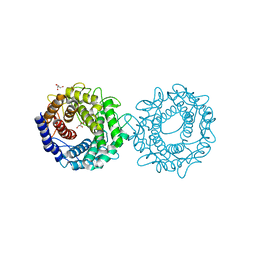 | | Crystal structure of glucose-2-epimerase from Runella slithyformis Runsl_4512 | | Descriptor: | FORMIC ACID, GLYCEROL, N-acylglucosamine 2-epimerase | | Authors: | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H1M
 
 | | Crystal structure of glucose-2-epimerase mutant_D254A from Runella slithyformis Runsl_4512 | | Descriptor: | FORMIC ACID, N-acylglucosamine 2-epimerase | | Authors: | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H1N
 
 | | Crystal structure of glucose-2-epimerase mutant_D254A in complex with D-Glucitol from Runella slithyformis Runsl_4512 | | Descriptor: | FORMIC ACID, N-acylglucosamine 2-epimerase, sorbitol | | Authors: | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7CN8
 
 | | Cryo-EM structure of PCoV_GX spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein, ... | | Authors: | Wang, X, Yu, J, Zhang, S, Qiao, S, Zeng, J, Tian, L. | | Deposit date: | 2020-07-30 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution.
Nat Commun, 12, 2021
|
|
7CN4
 
 | | Cryo-EM structure of bat RaTG13 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Zhang, S, Qiao, S, Yu, J, Zeng, J, Tian, L. | | Deposit date: | 2020-07-30 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution.
Nat Commun, 12, 2021
|
|
8IBK
 
 | | Crystal structure of Bacillus sp. AHU2216 GH13_31 Alpha-glucosidase E256Q/N258G in complex with maltotriose | | Descriptor: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Auiewiriyanukul, W, Saburi, W, Yu, J, Kato, K, Yao, M, Mori, H. | | Deposit date: | 2023-02-10 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Alteration of Substrate Specificity and Transglucosylation Activity of GH13_31 alpha-Glucosidase from Bacillus sp. AHU2216 through Site-Directed Mutagenesis of Asn258 on beta → alpha Loop 5.
Molecules, 28, 2023
|
|
8I3J
 
 | | Crystal structure of human inner-arm dynein heavy chain d stalk and microtubule binding domain | | Descriptor: | Dynein axonemal heavy chain 1 | | Authors: | Ko, S, Yu, J, Toda, A, Tanaka, H, Kurisu, G. | | Deposit date: | 2023-01-17 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of the stalk region of axonemal inner-arm dynein-d reveals unique features in the coiled-coil and microtubule-binding domain.
Febs Lett., 597, 2023
|
|
7E9X
 
 | | Trehalase of Arabidopsis thaliana acid mutant -D380A | | Descriptor: | GLYCEROL, Trehalase | | Authors: | Taguchi, Y, Saburi, W, Yu, J, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2021-03-05 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | pH-dependent alteration of substrate specificity of plant trehalase and its molecular mechanism
To Be Published
|
|
7E9U
 
 | | Trehalase of Arabidopsis thaliana | | Descriptor: | GLYCEROL, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Taguchi, Y, Saburi, W, Yu, J, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2021-03-05 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | pH-dependent alteration of substrate specificity of plant trehalase and its molecular mechanism
To Be Published
|
|
7EAW
 
 | | Trehalase of Arabidopsis thaliana acid mutant -D380A trehalose complex | | Descriptor: | GLYCEROL, Trehalase, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Taguchi, Y, Saburi, W, Yu, J, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2021-03-08 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | pH-dependent alteration of substrate specificity of plant trehalase and its molecular mechanism
To Be Published
|
|
8IN4
 
 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai | | Descriptor: | 25 kDa polyphenol-binding protein, ACETYL GROUP, GLYCEROL | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN1
 
 | | beta-glucosidase protein from Aplysia kurodai | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-Glucosidase, alpha-L-fucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN3
 
 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai | | Descriptor: | 25 kDa polyphenol-binding protein, GLYCEROL | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN6
 
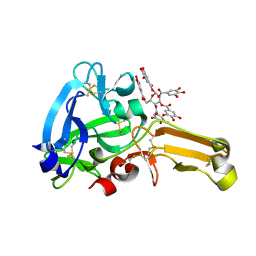 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai complex with tannic acid | | Descriptor: | 25 kDa polyphenol-binding protein, BETA-1,2,3,4,6-PENTA-O-GALLOYL-D-GLUCOPYRANOSE | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
