7WRI
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike receptor-binding domain in complex with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Han, P, Xie, Y, Qi, J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Broader-species receptor binding and structural bases of Omicron SARS-CoV-2 to both mouse and palm-civet ACE2s.
Cell Discov, 8, 2022
|
|
1MKS
 
 | |
7WRH
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Han, P, Xie, Y, Qi, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Broader-species receptor binding and structural bases of Omicron SARS-CoV-2 to both mouse and palm-civet ACE2s.
Cell Discov, 8, 2022
|
|
1MKU
 
 | |
7C6S
 
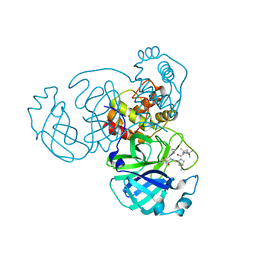 | |
7C6U
 
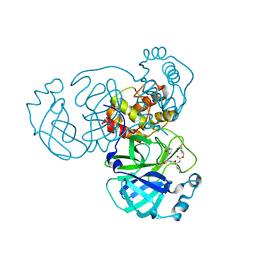 | | Crystal structure of SARS-CoV-2 complexed with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Fu, L, Feng, Y, Qi, J, Gao, F.G. | | Deposit date: | 2020-05-22 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease.
Nat Commun, 11, 2020
|
|
7D1M
 
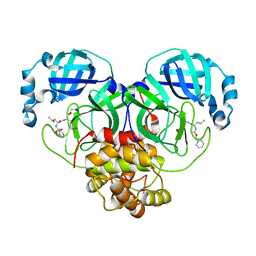 | | CRYSTAL STRUCTURE OF THE SARS-CoV-2 MAIN PROTEASE COMPLEXED WITH GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fu, L.F, Gilski, M, Shabalin, I, Gao, G.F, Qi, J.X. | | Deposit date: | 2020-09-14 | | Release date: | 2020-10-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease.
Nat Commun, 11, 2020
|
|
7EIN
 
 | | SARS-CoV-2 main proteinase complex with microbial metabolite leupeptin | | Descriptor: | 3C-like proteinase, leupeptin | | Authors: | Fu, L.F, Feng, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-03-31 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Microbial Metabolite Leupeptin in the Treatment of COVID-19 by Traditional Chinese Medicine Herbs.
Mbio, 12, 2021
|
|
1UNE
 
 | |
7KJS
 
 | | Crystal structure of CDK2/cyclin E in complex with PF-06873600 | | Descriptor: | 6-(difluoromethyl)-8-[(1R,2R)-2-hydroxy-2-methylcyclopentyl]-2-{[1-(methylsulfonyl)piperidin-4-yl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | McTigue, M.A, He, Y, Ferre, R.A. | | Deposit date: | 2020-10-26 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Discovery of PF-06873600, a CDK2/4/6 Inhibitor for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
1MKV
 
 | |
1MKT
 
 | |
1KVX
 
 | |
1SHC
 
 | | SHC PTB DOMAIN COMPLEXED WITH A TRKA RECEPTOR PHOSPHOPEPTIDE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | SHC, TRKA RECEPTOR PHOSPHOPEPTIDE | | Authors: | Zhou, M.-M, Ravichandran, K.S, Olejniczak, E.T, Petros, A.M, Meadows, R.P, Sattler, M, Harlan, J.E, Wade, W.S, Burakoff, S.J, Fesik, S.W. | | Deposit date: | 1996-03-27 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure and ligand recognition of the phosphotyrosine binding domain of Shc.
Nature, 378, 1995
|
|
7WBQ
 
 | |
7WBP
 
 | |
7WBL
 
 | | Cryo-EM structure of human ACE2 complexed with SARS-CoV-2 Omicron RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2021-12-17 | | Release date: | 2022-01-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2.
Cell, 185, 2022
|
|
7WNM
 
 | | Structure of SARS-CoV-2 Gamma variant receptor-binding domain complexed with high affinity human ACE2 mutant (T27F,R273Q) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Ma, R.Y, Han, P.C, Wang, Q.H, Han, P. | | Deposit date: | 2022-01-18 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A binding-enhanced but enzymatic activity-eliminated human ACE2 efficiently neutralizes SARS-CoV-2 variants.
Signal Transduct Target Ther, 7, 2022
|
|
7X7D
 
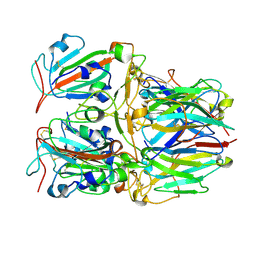 | | SARS-CoV-2 Delta RBD and Nb22 | | Descriptor: | Nb22, Spike protein S1 | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-03-09 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Short-Term Instantaneous Prophylaxis and Efficient Treatment Against SARS-CoV-2 in hACE2 Mice Conferred by an Intranasal Nanobody (Nb22).
Front Immunol, 13, 2022
|
|
7X7E
 
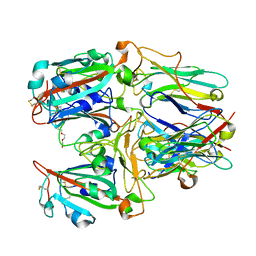 | | SARS-CoV-2 RBD and Nb22 | | Descriptor: | Nb22, Spike protein S1, TETRAETHYLENE GLYCOL | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-03-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Short-Term Instantaneous Prophylaxis and Efficient Treatment Against SARS-CoV-2 in hACE2 Mice Conferred by an Intranasal Nanobody (Nb22).
Front Immunol, 13, 2022
|
|
3QZ0
 
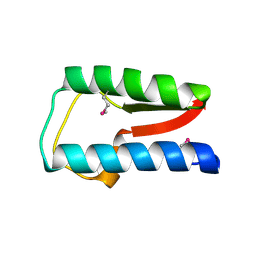 | | Structure of Treponema denticola Factor H Binding protein (FhbB), selenomethionine derivative | | Descriptor: | Factor H binding protein, GLYCEROL, THIOCYANATE ION | | Authors: | Miller, D.P, McDowell, J.V, Heroux, A, Bell, J.K, Marconi, R.T, Conrad, D.H, Burgner, J.W. | | Deposit date: | 2011-03-04 | | Release date: | 2012-03-07 | | Last modified: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of factor H-binding protein B (FhbB) of the periopathogen, Treponema denticola: insights into progression of periodontal disease.
J.Biol.Chem., 287, 2012
|
|
3R15
 
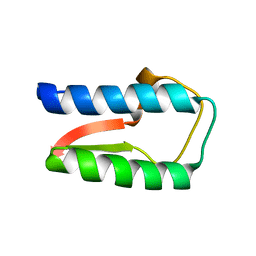 | | Structure Treponema Denticola Factor H Binding Protein | | Descriptor: | Factor H binding protein, THIOCYANATE ION | | Authors: | Miller, D.P, McDowell, J.V, Burgner, J, Heroux, A, Bell, J.K, Marconi, R.T. | | Deposit date: | 2011-03-09 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structure Treponema Denticola Factor H Binding Protein
To be Published
|
|
7K6M
 
 | | Crystal structure of PI3Kalpha selective Inhibitor PF-06843195 | | Descriptor: | 2,2-difluoroethyl (3S)-3-{[2'-amino-5-fluoro-2-(morpholin-4-yl)[4,5'-bipyrimidin]-6-yl]amino}-3-(hydroxymethyl)pyrrolidine-1-carboxylate, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Chen, P, Brooun, A, Deng, Y.L, Grodsky, N, Kaiser, S.E. | | Deposit date: | 2020-09-21 | | Release date: | 2021-01-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structure-Based Drug Design and Synthesis of PI3K alpha-Selective Inhibitor (PF-06843195).
J.Med.Chem., 64, 2021
|
|
7K6O
 
 | | Crystal structure of PI3Kalpha inhibitor 10-5429 | | Descriptor: | (3S)-3-[4-(2-aminopyrimidin-5-yl)-2-(morpholin-4-yl)-5,6-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl]-N-methylpyrrolidine-1-sulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Chen, P, Brooun, A, Deng, Y.L, Grodsky, N, Kaiser, S.E. | | Deposit date: | 2020-09-21 | | Release date: | 2021-01-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.738 Å) | | Cite: | Structure-Based Drug Design and Synthesis of PI3K alpha-Selective Inhibitor (PF-06843195).
J.Med.Chem., 64, 2021
|
|
7K6N
 
 | | Crystal structure of PI3Kalpha selective Inhibitor 11-1575 | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, tert-butyl (3S)-3-[4-(2-aminopyrimidin-5-yl)-2-(morpholin-4-yl)-5,6-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl]-3-methylpyrrolidine-1-carboxylate | | Authors: | Chen, P, Brooun, A, Deng, Y.L, Grodsky, N, Kaiser, S.E. | | Deposit date: | 2020-09-21 | | Release date: | 2021-01-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure-Based Drug Design and Synthesis of PI3K alpha-Selective Inhibitor (PF-06843195).
J.Med.Chem., 64, 2021
|
|
