1LP7
 
 | |
2Y1J
 
 | | CRYSTAL STRUCTURE OF A R-DIASTEREOMER ANALOGUE OF THE SPORE PHOTOPRODUCT IN COMPLEX WITH FRAGMENT DNA POLYMERASE I FROM BACILLUS STEAROTHERMOPHILUS | | Descriptor: | 5'-D(*AP*GP*GP*GP*QBTP*THM*GP*GP*TP*CP)-3', 5'-D(*GP*AP*CP*CP*AP*AP*CP*CP*CP*TP)-3', DNA POLYMERASE I, ... | | Authors: | Heil, K, Schneider, S, Mueller, M, Kneuttinger, A.C, Carell, T. | | Deposit date: | 2010-12-08 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures and Repair Studies Reveal the Identity and the Base-Pairing Properties of the Uv-Induced Spore Photoproduct DNA Lesion.
Chemistry, 17, 2011
|
|
4B9N
 
 | | Structure of the high fidelity DNA polymerase I correctly bypassing the oxidative formamidopyrimidine-dA DNA lesion. | | Descriptor: | 5'-D(*CP*AP*AP*(FAX)*AP*GP*AP*GP*TP*CP*AP*GP*GP*CP*TP)-3', 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*TP*TP*TP*TP)-3', DNA POLYMERASE, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
4B9L
 
 | | Structure of the high fidelity DNA polymerase I with the oxidative formamidopyrimidine-dA DNA lesion in the pre-insertion site. | | Descriptor: | 5'-D(*CP*AP*GP*FAX*AP*GP*AP*GP*TP*CP*AP*GP*GP*CP*TP)-3', 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*TP)-3', DNA POLYMERASE I, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
1M6F
 
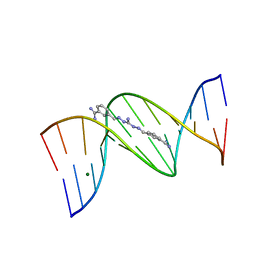 | | Strong Binding in the DNA Minor Groove by an Aromatic Diamidine With a Shape That Does Not Match the Curvature of the Groove | | Descriptor: | 3-[C-[N'-(3-CARBAMIMIDOYL-BENZYLIDENIUM)-HYDRAZINO]-[[AMINOMETHYLIDENE]AMINIUM]-IMINOMETHYL]-BENZAMIDINIUM, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Nguyen, B, Lee, M.P.H, Hamelberg, D, Joubert, A, Bailly, C, Brun, R, Neidle, S, Wilson, W.D. | | Deposit date: | 2002-07-16 | | Release date: | 2002-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Strong Binding in the DNA Minor Groove by an Aromatic Diamidine With a Shape That Does Not Match the Curvature of the Groove
J.Am.Chem.Soc., 124, 2002
|
|
4B9V
 
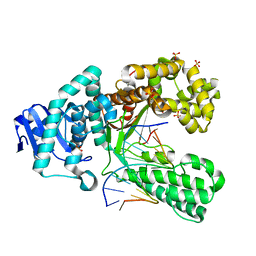 | | Structure of the high fidelity DNA polymerase I with extending from an oxidative formamidopyrimidine-dG DNA lesion -dA basepair. | | Descriptor: | 5'-D(*CP*AP*TP*FOXP*AP*GP*AP*GP*TP*CP*AP*GP*GP*CP*TP)-3', 5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*TP*AP*AP)-3', DNA POLYMERASE, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
4B9S
 
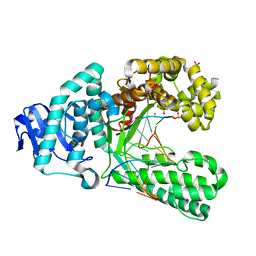 | | Structure of the high fidelity DNA polymerase I with an oxidative formamidopyrimidine-dG DNA lesion outside of the pre-insertion site. | | Descriptor: | 5'-D(*AP*CP*CP*TP*GP*AP*CP*TP*CP*TP)-3', 5'-D(*CP*AP*TP*FOXP*AP*GP*AP*GP*TP*CP*AP*GP*GP*TP*TP)-3', DNA POLYMERASE, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
3EM2
 
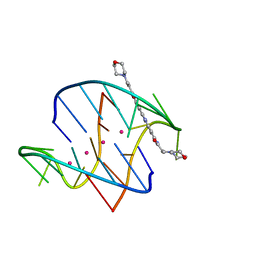 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6038 | | Descriptor: | 3,6-Bis[(3-morpholinopropionamido)] acridine, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-09-23 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
3EQW
 
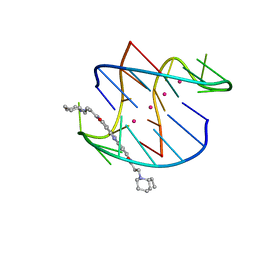 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6042 in small unit cell | | Descriptor: | 3,6-Bis{3-[(2R)-(2-ethylpiperidino)]propionamido}acridine, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
303D
 
 | | META-HYDROXY ANALOGUE OF HOECHST 33258 ('HYDROXYL OUT' CONFORMATION) BOUND TO D(CGCGAATTCGCG)2 | | Descriptor: | 3-[5-[5-(4-METHYL-PIPERAZIN-1-YL)-1H-IMIDAZO[4,5-B]PYRIDIN-2-YL]-BENZIMIDAZOL-2-YL]-PHENOL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Clark, G.R, Squire, C.J, Gray, E.J, Leupin, W, Neidle, S. | | Deposit date: | 1996-06-26 | | Release date: | 1997-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Designer DNA-binding drugs: the crystal structure of a meta-hydroxy analogue of Hoechst 33258 bound to d(CGCGAATTCGCG)2.
Nucleic Acids Res., 24, 1996
|
|
360D
 
 | | STRUCTURE OF 2,5-BIS{[4-(N-ETHYLAMIDINO)PHENYL]}FURAN COMPLEXED TO 5'-D(CPGPCPGPAPAPTPTPCPGPCPG)-3'. A MINOR GROOVE DRUG COMPLEX, SHOWING PATTERNS OF GROOVE HYDRATION | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION, N,N'-(furan-2,5-diylbis{benzene-4,1-diyl[(Z)-aminomethylylidene]})diethanaminium | | Authors: | Guerri, A, Simpson, I.J, Neidle, S. | | Deposit date: | 1997-10-29 | | Release date: | 1998-07-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Visualisation of extensive water ribbons and networks in a DNA minor-groove drug complex.
Nucleic Acids Res., 26, 1998
|
|
3ET8
 
 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6054 | | Descriptor: | 3,6-Bis{3-(3-[(3R)-methylpiperidino)]propionamido}acridine, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-07 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
311D
 
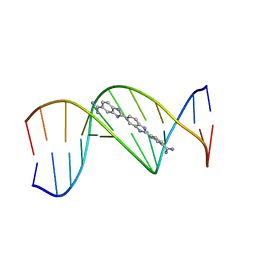 | | THE ROLE OF HYDROGEN BONDING IN MINOR-GROOVE DRUG-DNA RECOGNITION. STRUCTURE OF A BIS-AMIDINIUM DERIVATIVE OF HOECHST 33258 COMPLEXED TO THE DODECANUCLEOTIDE D(CGCGAATTCGCG)2 | | Descriptor: | 5-AMIDINO-2-[2-(4-AMIDINOPHENYL)-5-BENZIMIDAZOLYL]BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Clark, G.R, Boykin, D.W, Czarny, A, Neidle, S. | | Deposit date: | 1997-02-04 | | Release date: | 1997-02-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a bis-amidinium derivative of hoechst 33258 complexed to dodecanucleotide d(CGCGAATTCGCG)2: the role of hydrogen bonding in minor groove drug-DNA recognition.
Nucleic Acids Res., 25, 1997
|
|
302D
 
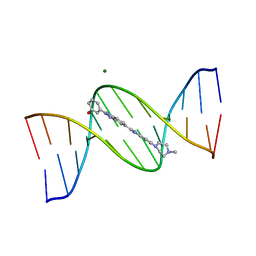 | | META-HYDROXY ANALOGUE OF HOECHST 33258 ('HYDROXYL IN' CONFORMATION) BOUND TO D(CGCGAATTCGCG)2 | | Descriptor: | 3-[5-[5-(4-METHYL-PIPERAZIN-1-YL)-1H-IMIDAZO[4,5-B]PYRIDIN-2-YL]-BENZIMIDAZOL-2-YL]-PHENOL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Clark, G.R, Squire, C.J, Gray, E.J, Leupin, W, Neidle, S. | | Deposit date: | 1996-06-26 | | Release date: | 1997-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Designer DNA-binding drugs: the crystal structure of a meta-hydroxy analogue of Hoechst 33258 bound to d(CGCGAATTCGCG)2.
Nucleic Acids Res., 24, 1996
|
|
335D
 
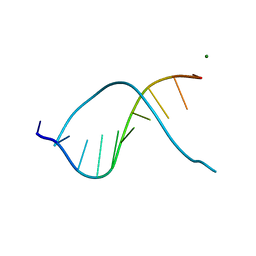 | |
2NLM
 
 | |
1OON
 
 | | Nitroreductase from e-coli in complex with the dinitrobenzamide prodrug SN27217 | | Descriptor: | 2,4-DINITRO,5-[BIS(2-BROMOETHYL)AMINO]-N-(2',3'-DIOXOPROPYL)BENZAMIDE, FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Johansson, E, Parkinson, G.N, Denny, W.A, Neidle, S. | | Deposit date: | 2003-03-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Studies on the Nitroreductase Prodrug-Activating System. Crystal Structures of Complexes with the Inhibitor Dicoumarol and Dinitrobenzamide Prodrugs and of the Enzyme Active Form
J.Med.Chem., 46, 2003
|
|
1OOQ
 
 | | Nitroreductase from e-coli in complex with the inhibitor dicoumarol | | Descriptor: | BISHYDROXY[2H-1-BENZOPYRAN-2-ONE,1,2-BENZOPYRONE], FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Johansson, E, Parkinson, G.N, Denny, W.A, Neidle, S. | | Deposit date: | 2003-03-04 | | Release date: | 2003-04-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies on the Nitroreductase Prodrug-Activating System. Crystal Structures of Complexes with the Inhibitor Dicoumarol and Dinitrobenzamide Prodrugs and of the Enzyme Active Form
J.Med.Chem., 46, 2003
|
|
2O6P
 
 | | Crystal Structure of the heme-IsdC complex | | Descriptor: | CHLORIDE ION, Iron-regulated surface determinant protein C, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Sharp, K.H, Schneider, S, Cockayne, A, Paoli, M. | | Deposit date: | 2006-12-08 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the heme-IsdC complex, the central conduit of the Isd iron/heme uptake system in Staphylococcus aureus.
J. Biol. Chem., 282, 2007
|
|
3EUI
 
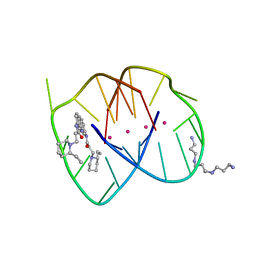 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6042 in a large unit cell | | Descriptor: | 3,6-Bis{3-[(2R)-(2-ethylpiperidino)]propionamido}acridine, 3-[(2R)-2-ethylpiperidin-1-yl]-N-[6-({3-[(2S)-2-ethylpiperidin-1-yl]propanoyl}amino)acridin-3-yl]propanamide, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', ... | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-10 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
3ERU
 
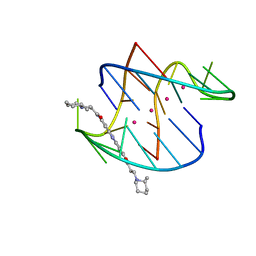 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6045 | | Descriptor: | 3,6-Bis{3-[(2R)-2-methylpiperidino)]propionamido}acridine, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-03 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
3EUM
 
 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6066 | | Descriptor: | 3,6-Bis[3-(azepan-1-yl)propionamido]acridine, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-10 | | Release date: | 2008-10-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
3ES0
 
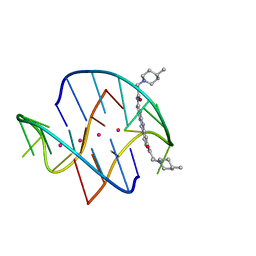 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6048 | | Descriptor: | 3,6-Bis[3-(4-methylpiperidino)propionamido]acridine, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-03 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
3CDM
 
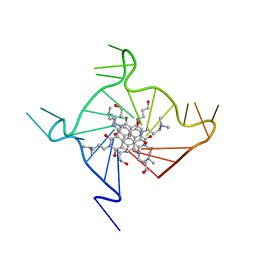 | | Structural adaptation and conservation in quadruplex-drug recognition | | Descriptor: | 2,7-bis[3-(dimethylamino)propyl]-4,9-bis[(3-hydroxypropyl)amino]benzo[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetrone, DNA (5'-D(*DT*DAP*DGP*DGP*DGP*DTP*DTP*DAP*DGP*DGP*DGP*DTP*DTP*DAP*DGP*DGP*DGP*DTP*DTP*DAP*DGP*DGP*DG)-3'), POTASSIUM ION | | Authors: | Parkinson, G.N, Neidle, S. | | Deposit date: | 2008-02-27 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Topology conservation and loop flexibility in quadruplex-drug recognition: crystal structures of inter- and intramolecular telomeric DNA quadruplex-drug complexes
J.Mol.Biol., 381, 2008
|
|
3CE5
 
 | | A bimolecular parallel-stranded human telomeric quadruplex in complex with a 3,6,9-trisubstituted acridine molecule BRACO19 | | Descriptor: | 9-[4-(n,n-dimethylamino)phenylamino]-3,6-bis(3-pyrrolidinopropionamido) acridine, DNA (5'-D(*DTP*DAP*DGP*DGP*DGP*DTP*DTP*DAP*DGP*DGP*DGP*DT)-3'), POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G.N, Reszka, A.P, Neidle, S. | | Deposit date: | 2008-02-28 | | Release date: | 2008-05-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of DNA quadruplex recognition by an acridine drug.
J.Am.Chem.Soc., 130, 2008
|
|
