8BZ6
 
 | |
8BZ5
 
 | |
4PAZ
 
 | | OXIDIZED MUTANT P80A PSEUDOAZURIN FROM A. FAECALIS | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Adman, E.T, Libeu, C.A.P. | | Deposit date: | 1997-02-20 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Site-directed mutants of pseudoazurin: explanation of increased redox potentials from X-ray structures and from calculation of redox potential differences.
Biochemistry, 36, 1997
|
|
8TC9
 
 | | Human asparaginyl-tRNA synthetase bound to OSM-S-106 | | Descriptor: | Asparagine--tRNA ligase, cytoplasmic, GLYCEROL, ... | | Authors: | Dogovski, C, Metcalfe, R.D, Xie, S.C, Morton, C.J, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2023-06-30 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reaction hijacking inhibition of Plasmodium falciparum asparagine tRNA synthetase.
Nat Commun, 15, 2024
|
|
8TC8
 
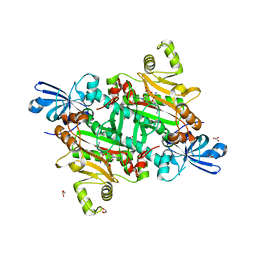 | | Human asparaginyl-tRNA synthetase bound to adenosine 5'-sulfamate | | Descriptor: | 5'-O-[N-(L-ASPARAGINYL)SULFAMOYL]ADENOSINE, Asparagine--tRNA ligase, cytoplasmic, ... | | Authors: | Dogovski, C, Metcalfe, R.D, Xie, S.C, Morton, C.J, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2023-06-30 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reaction hijacking inhibition of Plasmodium falciparum asparagine tRNA synthetase.
Nat Commun, 15, 2024
|
|
8TC7
 
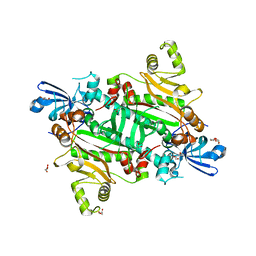 | | Human asparaginyl-tRNA synthetase, apo form | | Descriptor: | Asparagine--tRNA ligase, cytoplasmic, CHLORIDE ION, ... | | Authors: | Dogovski, C, Metcalfe, R.D, Xie, S.C, Morton, C.J, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2023-06-30 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reaction hijacking inhibition of Plasmodium falciparum asparagine tRNA synthetase.
Nat Commun, 15, 2024
|
|
8DKN
 
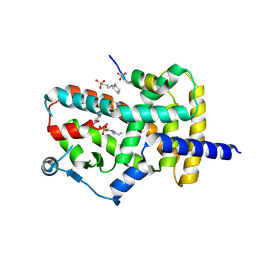 | | PPARg bound to T0070907 and Co-R peptide | | Descriptor: | 2-chloro-5-nitro-N-(pyridin-4-yl)benzamide, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Nuclear receptor corepressor 1 peptide, ... | | Authors: | Larsen, N.A, Tsai, J. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
8DSZ
 
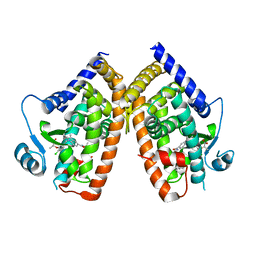 | | PPARg bound to partial agonist H3B-487 | | Descriptor: | (2R)-2-{5-[(5-{[(1R)-1-(4-tert-butylphenyl)ethyl]carbamoyl}-2,3-dimethyl-1H-indol-1-yl)methyl]-2-chlorophenoxy}propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Larsen, N.A. | | Deposit date: | 2022-07-24 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
4LE1
 
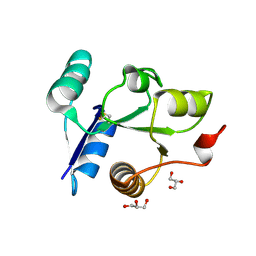 | | Crystal structure of the receiver domain of DesR in the inactive state | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Trajtenberg, F, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Allosteric activation of bacterial response regulators: the role of the cognate histidine kinase beyond phosphorylation.
MBio, 5, 2014
|
|
4LE2
 
 | |
5EDC
 
 | |
9GZJ
 
 | |
9GZK
 
 | |
6G54
 
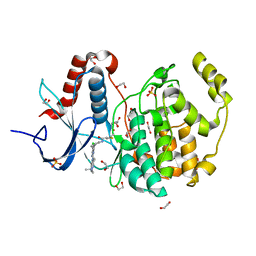 | | Crystal structure of ERK2 covalently bound to SM1-71 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Mitogen-activated protein kinase 1, ... | | Authors: | Chaikuad, A, Suman, R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-29 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Leveraging Compound Promiscuity to Identify Targetable Cysteines within the Kinome.
Cell Chem Biol, 26, 2019
|
|
6GES
 
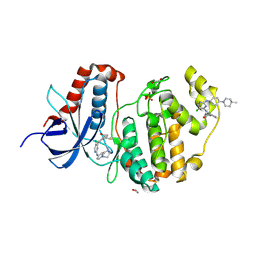 | | Crystal structure of ERK1 covalently bound to SM1-71 | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 3, N-{2-[(5-chloro-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]phenyl}propanamide, ... | | Authors: | Chaikuad, A, Suman, R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-27 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Leveraging Compound Promiscuity to Identify Targetable Cysteines within the Kinome.
Cell Chem Biol, 26, 2019
|
|
5EDG
 
 | |
5EDB
 
 | |
5EDH
 
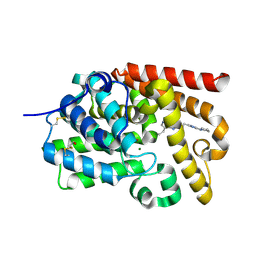 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n4c(C)n1c(nc(n1)CCc2nc(nn2C)N3CCCC3)c(c4)CC, micromolar IC50=0.0037753 | | Descriptor: | 8-ethyl-5-methyl-2-[2-(2-methyl-5-pyrrolidin-1-yl-1,2,4-triazol-3-yl)ethyl]-[1,2,4]triazolo[1,5-c]pyrimidine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Rudolph, M.G. | | Deposit date: | 2015-10-21 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
5EDE
 
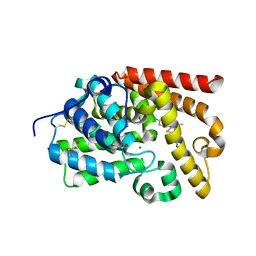 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c13c(cc(s1)C(NCC2OCCC2)=O)c(nn3c4ccc(cc4)Cl)C, micromolar IC50=0.217 | | Descriptor: | 1-(4-chlorophenyl)-3-methyl-~{N}-[[(2~{R})-oxolan-2-yl]methyl]thieno[2,3-c]pyrazole-5-carboxamide, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Joseph, C, Rudolph, M.G. | | Deposit date: | 2015-10-21 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
5EDI
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c2(c(n1nc(nc1c(c2)C)CCc3nc(nn3C)N4CCCC4)C)Cl, micromolar IC50=0.000279 | | Descriptor: | 6-chloranyl-5,8-dimethyl-2-[2-(2-methyl-5-pyrrolidin-1-yl-1,2,4-triazol-3-yl)ethyl]-[1,2,4]triazolo[1,5-a]pyridine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Rudolph, M.G. | | Deposit date: | 2015-10-21 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
6R5F
 
 | | Crystal structure of RIP1 kinase in complex with DHP77 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 1, [(5~{S})-5-[3,5-bis(fluoranyl)phenyl]pyrazolidin-1-yl]-[1-(5-methyl-1,3,4-oxadiazol-2-yl)piperidin-4-yl]methanone | | Authors: | Thorpe, J.H, Campobasso, N, Harris, P.A. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Discovery and Lead-Optimization of 4,5-Dihydropyrazoles as Mono-Kinase Selective, Orally Bioavailable and Efficacious Inhibitors of Receptor Interacting Protein 1 (RIP1) Kinase.
J.Med.Chem., 62, 2019
|
|
5PAZ
 
 | | REDUCED MUTANT P80A PSEUDOAZURIN FROM A. FAECALIS | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Adman, E.T, Libeu, C.A.P. | | Deposit date: | 1997-02-21 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Site-directed mutants of pseudoazurin: explanation of increased redox potentials from X-ray structures and from calculation of redox potential differences.
Biochemistry, 36, 1997
|
|
1R6H
 
 | | Solution Structure of human PRL-3 | | Descriptor: | protein tyrosine phosphatase type IVA, member 3 isoform 1 | | Authors: | Kozlov, G, Gehring, K, Ekiel, I. | | Deposit date: | 2003-10-15 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into Molecular Function of the Metastasis-associated Phosphatase PRL-3.
J.Biol.Chem., 279, 2004
|
|
1D5G
 
 | |
5NPH
 
 | |
