7F1D
 
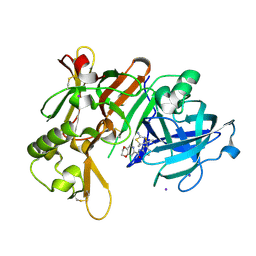 | | Crystal Structure of BACE1 in complex with N-{3-[(4R,5R,6R)-2-amino-5-fluoro-4,6-dimethyl-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-2H,3H-[1,4]dioxino[2,3-c]pyridine-7-carboxamide | | Descriptor: | Beta-secretase 1, IODIDE ION, N-[3-[(4R,5R,6R)-2-azanyl-5-fluoranyl-4,6-dimethyl-5,6-dihydro-1,3-thiazin-4-yl]-4-fluoranyl-phenyl]-2,3-dihydro-[1,4]dioxino[2,3-c]pyridine-7-carboxamide | | Authors: | Ueno, T, Matsuoka, E, Asada, N, Yamamoto, S, Kanegawa, N, Ito, M, Ito, H, Moechars, D, Rombouts, F.J.R, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2021-06-09 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Extremely Selective Fused Pyridine-Derived beta-Site Amyloid Precursor Protein-Cleaving Enzyme (BACE1) Inhibitors with High In Vivo Efficacy through 10s Loop Interactions.
J.Med.Chem., 64, 2021
|
|
9QLU
 
 | |
7F1G
 
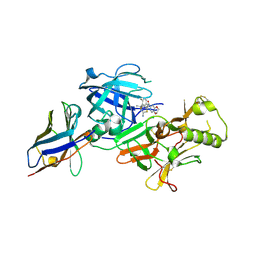 | | BACE2 xaperone complex with N-{3-[(4R,5R,6R)-2-amino-5-fluoro-4,6-dimethyl-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-2H,3H-[1,4]dioxino[2,3-c]pyridine-7-carboxamide | | Descriptor: | Beta-secretase 2, N-[3-[(4R,5R,6R)-2-azanyl-5-fluoranyl-4,6-dimethyl-5,6-dihydro-1,3-thiazin-4-yl]-4-fluoranyl-phenyl]-2,3-dihydro-[1,4]dioxino[2,3-c]pyridine-7-carboxamide, XAPERONE | | Authors: | Ueno, T, Matsuoka, E, Asada, N, Yamamoto, S, Kanegawa, N, Ito, M, Ito, H, Moechars, D, Rombouts, F.J.R, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2021-06-09 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Extremely Selective Fused Pyridine-Derived beta-Site Amyloid Precursor Protein-Cleaving Enzyme (BACE1) Inhibitors with High In Vivo Efficacy through 10s Loop Interactions.
J.Med.Chem., 64, 2021
|
|
7F4A
 
 | |
9UGE
 
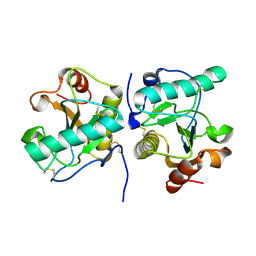 | | Crystal structure of the complex of camel peptidoglycan recognition protein, PGRP-S with malic acid and oxalic acid at 2.3 A resolution | | Descriptor: | D-MALATE, OXALIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Barik, D, Ahmad, N, Maurya, A, Yamini, S, Sharma, P, Yadav, S.P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2025-04-11 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | Crystal structure of the complex of camel peptidoglycan recognition protein, PGRP-S with malic acid and oxalic acid at 2.3 A resolution
To Be Published
|
|
7F2J
 
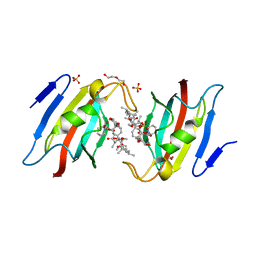 | | Crystal structure of AtFKBP53 FKBD in complex with rapamycin | | Descriptor: | DI(HYDROXYETHYL)ETHER, Peptidyl-prolyl cis-trans isomerase FKBP53, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, ... | | Authors: | Singh, A.K, Saharan, K, Vasudevan, D. | | Deposit date: | 2021-06-11 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal packing reveals rapamycin-mediated homodimerization of an FK506-binding domain.
Int.J.Biol.Macromol., 206, 2022
|
|
7F3S
 
 | |
7F51
 
 | | Crystal structure of Hst2 in complex with 2'-O-Benzoyl ADP Ribose | | Descriptor: | NAD-dependent protein deacetylase HST2, ZINC ION, [(2R,3R,4R,5R)-5-[[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-2,4-bis(oxidanyl)oxolan-3-yl] benzoate | | Authors: | Wang, D, Yan, F, Chen, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Global profiling of regulatory elements in the histone benzoylation pathway.
Nat Commun, 13, 2022
|
|
6M6Z
 
 | | A de novo designed transmembrane nanopore, TMH4C4 | | Descriptor: | TMH4C4 | | Authors: | Lu, P, Xu, C, Reggiano, G, Xu, Q, DiMaio, F, Baker, D. | | Deposit date: | 2020-03-16 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
7F5M
 
 | |
7F4E
 
 | |
7F86
 
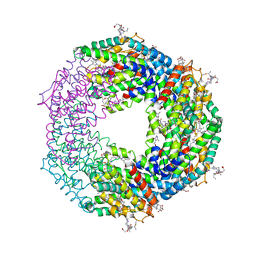 | | Crystal structure of Phycoerythrin from Halomicronema Sp. R31DM | | Descriptor: | PHYCOERYTHROBILIN, Phycoerythrin alpha subunit, Phycoerythrin beta subunit | | Authors: | Patel, S.N, Gupta, G.D, Sonani, R.R, Singh, N.K, Kumar, V, Madamwar, D. | | Deposit date: | 2021-07-01 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure analysis of phycoerythrin from marine cyanobacterium Halomicronema .
J.Biomol.Struct.Dyn., 41, 2023
|
|
7EZC
 
 | | Adenosine A2a receptor mutant-I92N | | Descriptor: | 6-(2,2-diphenylethylamino)-9-[(2R,3R,4S,5S)-5-(ethylcarbamoyl)-3,4-dihydroxy-oxolan-2-yl]-N-[2-[(1-pyridin-2-ylpiperidin-4-yl)carbamoylamino]ethyl]purine-2-carboxamide, Adenosine receptor A2a,Soluble cytochrome b562 | | Authors: | Cui, M, Zhou, Q, Yao, D, Zhao, S, Song, G. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of a constitutive active mutant of adenosine A 2A receptor.
Iucrj, 9, 2022
|
|
7F1U
 
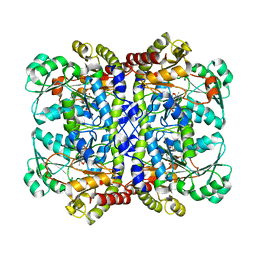 | | Crystal structure of Pseudomonas putida methionine gamma-lyase Q349S mutant with L-methionine intermediates | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-4-(methylsulfanyl)but-2-enoic acid, L-methionine gamma-lyase, METHIONINE | | Authors: | Okawa, A, Handa, H, Yasuda, E, Murota, M, Kudo, D, Tamura, T, Shiba, T, Inagaki, K. | | Deposit date: | 2021-06-09 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization and application of l-methionine gamma-lyase Q349S mutant enzyme with an enhanced activity toward l-homocysteine.
J.Biosci.Bioeng., 133, 2022
|
|
7F1P
 
 | | Crystal structure of Pseudomonas putida methionine gamma-lyase Q349S mutant ligand-free form. | | Descriptor: | L-methionine gamma-lyase | | Authors: | Okawa, A, Handa, H, Yasuda, E, Murota, M, Kudo, D, Tamura, T, Shiba, T, Inagaki, K. | | Deposit date: | 2021-06-09 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization and application of l-methionine gamma-lyase Q349S mutant enzyme with an enhanced activity toward l-homocysteine.
J.Biosci.Bioeng., 133, 2022
|
|
7F1V
 
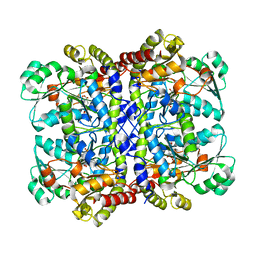 | | Crystal structure of Pseudomonas putida methionine gamma-lyase Q349S mutant with L-homocysteine intermediates | | Descriptor: | (2~{S})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-4-sulfanyl-butanoic acid, 2-AMINO-4-MERCAPTO-BUTYRIC ACID, L-methionine gamma-lyase | | Authors: | Okawa, A, Handa, H, Yasuda, E, Murota, M, Kudo, D, Tamura, T, Shiba, T, Inagaki, K. | | Deposit date: | 2021-06-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterization and application of l-methionine gamma-lyase Q349S mutant enzyme with an enhanced activity toward l-homocysteine.
J.Biosci.Bioeng., 133, 2022
|
|
7F30
 
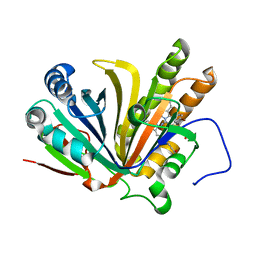 | | Crystal structure of OxdB E85A in complex with Z-2- (3-bromophenyl) propanal oxime | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Phenylacetaldoxime dehydratase, Z-2-(3-bromophenyl) propanal oxime | | Authors: | Muraki, N, Matsui, D, Asano, Y, Aono, S. | | Deposit date: | 2021-06-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structural analysis of aldoxime dehydratase from Bacillus sp. OxB-1: Importance of surface residues in optimization for crystallization.
J.Inorg.Biochem., 230, 2022
|
|
7F2Y
 
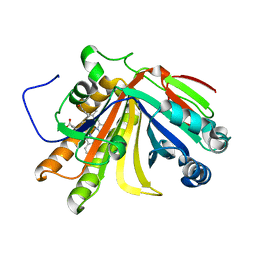 | | Crystal structure of OxdB E85A mutant (form I) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Phenylacetaldoxime dehydratase | | Authors: | Muraki, N, Matsui, D, Asano, Y, Aono, S. | | Deposit date: | 2021-06-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structural analysis of aldoxime dehydratase from Bacillus sp. OxB-1: Importance of surface residues in optimization for crystallization.
J.Inorg.Biochem., 230, 2022
|
|
7F2Z
 
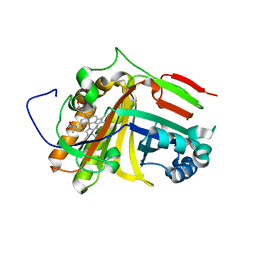 | | Crystal structure of OxdB E85A mutant (form II) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Phenylacetaldoxime dehydratase | | Authors: | Muraki, N, Matsui, D, Asano, Y, Aono, S. | | Deposit date: | 2021-06-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structural analysis of aldoxime dehydratase from Bacillus sp. OxB-1: Importance of surface residues in optimization for crystallization.
J.Inorg.Biochem., 230, 2022
|
|
7ERL
 
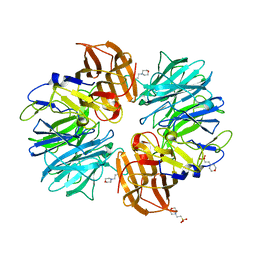 | |
7ES4
 
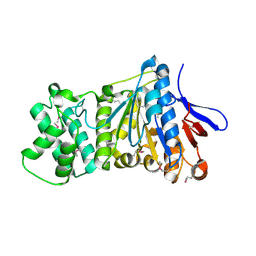 | | the crystral structure of DndH-C-domain | | Descriptor: | DNA phosphorothioation-dependent restriction protein DptH | | Authors: | Wu, D. | | Deposit date: | 2021-05-08 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The functional coupling between restriction and DNA phosphorothioate modification systems underlying the DndFGH restriction complex
Nat Catal, 2022
|
|
7FAR
 
 | | Crystal structure of PDE5A in complex with inhibitor L12 | | Descriptor: | 5-[bis(fluoranyl)methoxy]-2-[(4-chlorophenyl)methyl]-10-(3-methoxypropyl)-3,10-diazatricyclo[6.4.1.0^{4,13}]trideca-1,4(13),5,7-tetraen-9-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wu, D, Huang, Y.Y, Luo, H.B. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.40006471 Å) | | Cite: | Free energy perturbation (FEP)-guided scaffold hopping.
Acta Pharm Sin B, 12, 2022
|
|
7FB8
 
 | | De Novo-Designed and Disulfide-Bridged Peptide Heterodimer - hd1 | | Descriptor: | ASP-ASP-LYS-ASP-CYS-ASP-GLU-TYR-CYS-LYS-LYS-THR-LYS-GLU-NH2, GLU-LE1-THR-GLY-HIS-ILE-GLU-GLY-PRO-THR-LE1-THR-LE1-HIS-CYS-LYS-NH2 | | Authors: | Yao, H, Yao, S, Zheng, Y, Moyer, A, Baker, D, Wu, C. | | Deposit date: | 2021-07-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | De novo design and directed folding of disulfide-bridged peptide heterodimers.
Nat Commun, 13, 2022
|
|
7FBA
 
 | | De Novo-Designed and Disulfide-Bridged Peptide Heterodimer - hd2 | | Descriptor: | ALA-LE1-CYS-GLU-CYS-GLY-PRO-THR-ARG-GLU-CYS-LYS-NH2, GLU-CYS-ARG-GLU-TYR-GLY-PRO-LE1-LYS-LE1-LE1-ALA-NH2 | | Authors: | Yao, H, Yao, S, Moyer, A, Zheng, Y, Baker, D, Wu, C. | | Deposit date: | 2021-07-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | De novo design and directed folding of disulfide-bridged peptide heterodimers.
Nat Commun, 13, 2022
|
|
7ESH
 
 | | Crystal structure of amylosucrase from Calidithermus timidus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, amylosucrase | | Authors: | Tian, Y, Hou, X, Ni, D, Xu, W, Guang, C, Zhang, W, Rao, Y, Mu, W. | | Deposit date: | 2021-05-10 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure-based interface engineering methodology in designing a thermostable amylose-forming transglucosylase
J.Biol.Chem., 298, 2022
|
|
