4MOT
 
 | | Structure of Streptococcus pneumonia pare in complex with AZ13072886 | | Descriptor: | 1-[4-(3-methylbutyl)-5-oxo-6-(pyridin-3-yl)-4,5-dihydro[1,3]thiazolo[5,4-b]pyridin-2-yl]-3-prop-2-en-1-ylurea, Topoisomerase IV subunit B | | Authors: | Ogg, D, Boriack-Sjodin, P.A. | | Deposit date: | 2013-09-12 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thiazolopyridone ureas as DNA gyrase B inhibitors: Optimization of antitubercular activity and efficacy.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4AKX
 
 | | Structure of the heterodimeric complex ExoU-SpcU from the type III secretion system (T3SS) of Pseudomonas aeruginosa | | Descriptor: | EXOU, SPCU | | Authors: | Gendrin, C, Contreras-Martel, C, Bouillot, S, Elsen, S, Lemaire, D, Skoufias, D.A, Huber, P, Attree, I, Dessen, A. | | Deposit date: | 2012-02-29 | | Release date: | 2012-04-18 | | Last modified: | 2012-05-30 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structural Basis of Cytotoxicity Mediated by the Type III Secretion Toxin Exou from Pseudomonas Aeruginosa
Plos Pathog., 8, 2012
|
|
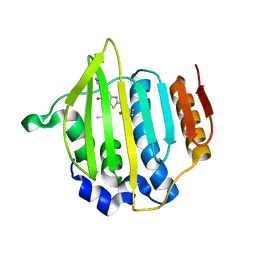
3CS8
 
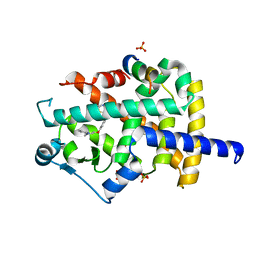 | |
8A6L
 
 | |
1EUB
 
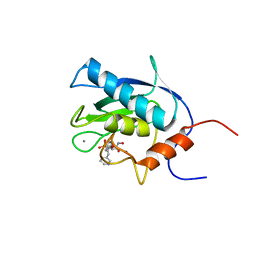 | | SOLUTION STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN COLLAGENASE-3 (MMP-13) COMPLEXED TO A POTENT NON-PEPTIDIC SULFONAMIDE INHIBITOR | | Descriptor: | 1-METHYLOXY-4-SULFONE-BENZENE, 3-METHYLPYRIDINE, CALCIUM ION, ... | | Authors: | Zhang, X, Gonnella, N.C, Koehn, J, Pathak, N, Ganu, V, Melton, R, Parker, D, Hu, S.I, Nam, K.Y. | | Deposit date: | 2000-04-14 | | Release date: | 2001-04-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human collagenase-3 (MMP-13) complexed to a potent non-peptidic sulfonamide inhibitor: binding comparison with stromelysin-1 and collagenase-1.
J.Mol.Biol., 301, 2000
|
|
7ZR2
 
 | | Crystal structure of a chimeric protein mimic of SARS-CoV-2 Spike HR1 in complex with HR2 | | Descriptor: | Spike protein S2', Spike protein S2',Chimeric protein mimic of SARS-CoV-2 Spike HR1 | | Authors: | Camara-Artigas, A, Gavira, J.A, Cano-Munoz, M, Polo-Megias, D, Conejero-Lara, F. | | Deposit date: | 2022-05-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel chimeric proteins mimicking SARS-CoV-2 spike epitopes with broad inhibitory activity.
Int.J.Biol.Macromol., 222, 2022
|
|
2OJ1
 
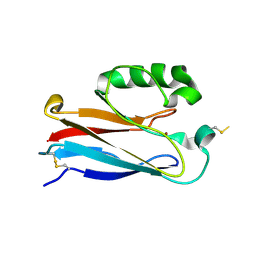 | | Disulfide-linked dimer of azurin N42C/M64E double mutant | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | de Jongh, T.E, Hoffmann, M, Einsle, O, Cavazzini, D, Rossi, G.L, Ubbink, M, Canters, G.W. | | Deposit date: | 2007-01-12 | | Release date: | 2007-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inter- and intramolecular electron transfer in modified azurin dimers
Eur.J.Inorg.Chem., 2007, 2007
|
|
7ZRD
 
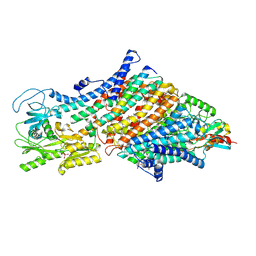 | | Cryo-EM map of the WT KdpFABC complex in the E1-P tight conformation, stabilised with the inhibitor orthovanadate | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
8F0V
 
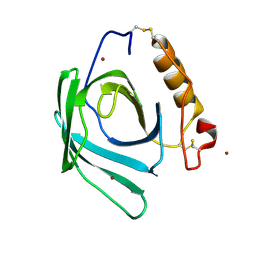 | | Lipocalin-like Milk protein-2 - E38A mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Milk protein, ZINC ION | | Authors: | Subramanian, R, KanagaVijayan, D. | | Deposit date: | 2022-11-04 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Variability in phenylalanine side chain conformations facilitates broad substrate tolerance of fatty acid binding in cockroach milk proteins.
Plos One, 18, 2023
|
|
8F0Y
 
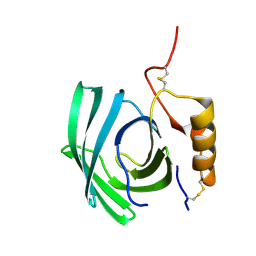 | | Lipocalin-like Milk protein-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Milk protein | | Authors: | Subramanian, R, KanagaVijayan, D, Shantakumar, R.P.S. | | Deposit date: | 2022-11-04 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Variability in phenylalanine side chain conformations facilitates broad substrate tolerance of fatty acid binding in cockroach milk proteins.
Plos One, 18, 2023
|
|
2OKE
 
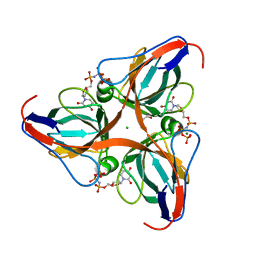 | | High Resolution Crystal Structures of Vaccinia Virus dUTPase | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2007-01-16 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of vaccinia virus dUTPase and its nucleotide complexes.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
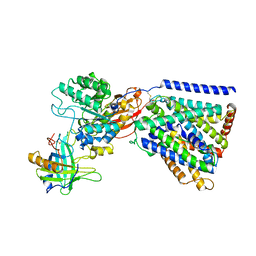
3JRG
 
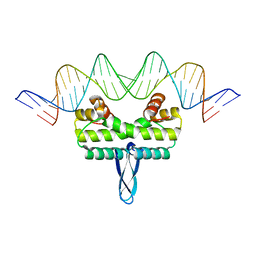 | |
2NUD
 
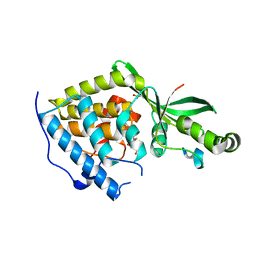 | | The structure of the type III effector AvrB complexed with a high-affinity RIN4 peptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Avirulence B protein, RPM1-interacting protein 4, ... | | Authors: | Singer, A.U, Desveaux, D, Wu, A.J, McNulty, B, Sondek, J, Dangl, J.L. | | Deposit date: | 2006-11-09 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Type III Effector Activation via Nucleotide Binding, Phosphorylation, and Host Target Interaction.
Plos Pathog., 3, 2007
|
|
3JRW
 
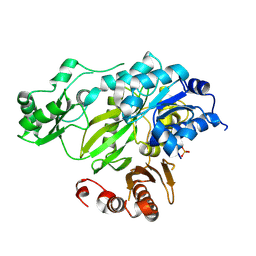 | | Phosphorylated BC domain of ACC2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | Deposit date: | 2009-09-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism for the regulation of human ACC2 through phosphorylation by AMPK
Biochem.Biophys.Res.Commun., 391, 2010
|
|
3CXV
 
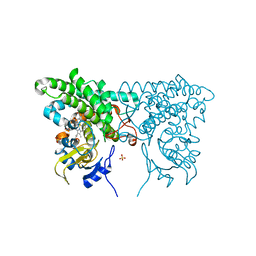 | |
3CXX
 
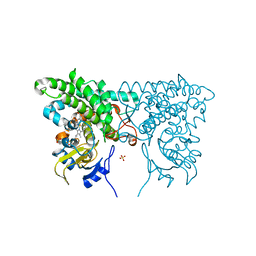 | |
2NVQ
 
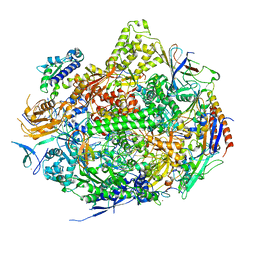 | | RNA Polymerase II Elongation Complex in 150 mM Mg+2 with 2'dUTP | | Descriptor: | 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', 5'-R(*AP*UP*CP*GP*AP*GP*AP*GP*GP*A)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-13 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
3ZKR
 
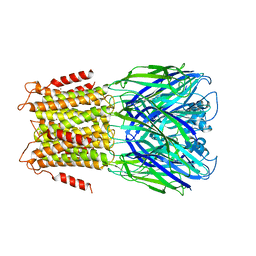 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with bromoform | | Descriptor: | CYS-LOOP LIGAND-GATED ION CHANNEL, TRIBROMOMETHANE | | Authors: | Spurny, R, Billen, B, Howard, R.J, Brams, M, Debaveye, S, Price, K.L, Weston, D.A, Strelkov, S.V, Tytgat, J, Bertrand, S, Bertrand, D, Lummis, S.C.R, Ulens, C. | | Deposit date: | 2013-01-24 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.649 Å) | | Cite: | Multisite Binding of a General Anesthetic to the Prokaryotic Pentameric Erwinia Chrysanthemi Ligand-Gated Ion Channel (Elic).
J.Biol.Chem., 288, 2013
|
|
6TM5
 
 | |
8AAC
 
 | |
2NWL
 
 | | Crystal structure of GltPh in complex with L-Asp | | Descriptor: | ASPARTIC ACID, PALMITIC ACID, glutamate symport protein | | Authors: | Gouaux, E, Boudker, O, Ryan, R, Yernool, D, Shimamoto, K. | | Deposit date: | 2006-11-15 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter.
Nature, 445, 2007
|
|
2NY1
 
 | | HIV-1 gp120 Envelope Glycoprotein (I109C, T257S, S334A, S375W, Q428C) Complexed with CD4 and Antibody 17b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, HEAVY CHAIN, ... | | Authors: | Zhou, T, Xu, L, Dey, B, Hessell, A.J, Van Ryk, D, Xiang, S.H, Yang, X, Zhang, M.Y, Zwick, M.B, Arthos, J, Burton, D.R, Dimitrov, D.S, Sodroski, J, Wyatt, R, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2006-11-20 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural definition of a conserved neutralization epitope on HIV-1 gp120.
Nature, 445, 2007
|
|
3JW8
 
 | | Crystal structure of human mono-glyceride lipase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, MGLL protein | | Authors: | Bertrand, T, Auge, F, Houtmann, J, Rak, A, Vallee, F, Mikol, V, Berne, P.F, Michot, N, Cheuret, D, Hoornaert, C, Mathieu, M. | | Deposit date: | 2009-09-18 | | Release date: | 2010-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for human monoglyceride lipase inhibition.
J.Mol.Biol., 396, 2010
|
|
3ZRB
 
 | | Structural basis for agonism and antagonism for a set of chemically related progesterone receptor modulators | | Descriptor: | (R)-N-[1-[4-(3,5-DIMETHYLISOXAZOL-4-YL)PHENYL]ETHYL]-3,5-DIMETHYLISOXAZOLE-4-SULFONAMIDE, 2-CHLORO-N-[[4-(3,5-DIMETHYLISOXAZOL-4-YL)PHENYL]METHYL]-1,4-DIMETHYL-1H-PYRAZOLE-4-SULFONAMIDE, GLYCEROL, ... | | Authors: | Lusher, S.J, Raaijmakers, H.C.A, Vu-Pham, D, Dechering, K, Wai Lam, T, Brown, A.R, Hamilton, N.M, Nimz, O, Azevedo, R, Mcguire, R, Oubrie, A, de Vlieg, J. | | Deposit date: | 2011-06-15 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for agonism and antagonism for a set of chemically related progesterone receptor modulators.
J. Biol. Chem., 286, 2011
|
|
3ZRH
 
 | | Crystal structure of the Lys29, Lys33-linkage-specific TRABID OTU deubiquitinase domain reveals an Ankyrin-repeat ubiquitin binding domain (AnkUBD) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, UBIQUITIN THIOESTERASE ZRANB1 | | Authors: | Licchesi, J.D.F, Akutsu, M, Komander, D. | | Deposit date: | 2011-06-16 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | An Ankyrin-Repeat Ubiquitin-Binding Domain Determines Trabid'S Specificity for Atypical Ubiquitin Chains.
Nat.Struct.Mol.Biol., 19, 2011
|
|
