7V9G
 
 | | Native BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(*GP*CP*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*CP*CP*A)-3'), DNA (5'-D(*TP*GP*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*TP*GP*C)-3') | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9I
 
 | | The Monomer mutant of BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*T)-3') | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9H
 
 | | The BEN3 domain of protein Bend3 | | Descriptor: | BEN domain-containing protein 3 | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9F
 
 | | Selenomethionine mutant (L740Sem) of BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, CITRIC ACID, DNA (5'-D(*GP*CP*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*CP*CP*A)-3'), ... | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7X7V
 
 | | Cryo-EM structure of SARS-CoV spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zhang, T, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X7T
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zheng, Q, Li, S, Zhang, T, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X7U
 
 | | Cryo-EM structure of SARS-CoV-2 Delta variant spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zhang, T, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8WNG
 
 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Ile | | Descriptor: | ACETATE ION, GLYCEROL, ISOLEUCINE, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-05 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8WO3
 
 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Mupirocin | | Descriptor: | ACETATE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8WNF
 
 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in apo form | | Descriptor: | ACETATE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-05 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8WNI
 
 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Val | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8WO2
 
 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Val-AMP | | Descriptor: | ACETATE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8WNJ
 
 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Ile-AMP | | Descriptor: | ACETATE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8IQX
 
 | | ferritin mutant-P156H | | Descriptor: | Ferritin | | Authors: | Zhao, G, Zhang, C, Zang, J, Zhang, T. | | Deposit date: | 2023-03-17 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Preparation and Unique Three-Dimensional Self-Assembly Property of Starfish Ferritin.
Foods, 12, 2023
|
|
8IR0
 
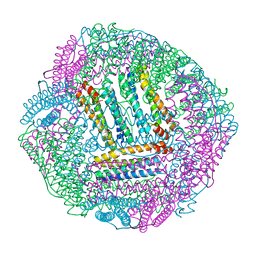 | | AfFer mutant-P156F | | Descriptor: | Ferritin | | Authors: | Zhao, G, Zhang, C, Zang, J, Zhang, T. | | Deposit date: | 2023-03-17 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Preparation and Unique Three-Dimensional Self-Assembly Property of Starfish Ferritin.
Foods, 12, 2023
|
|
8YLP
 
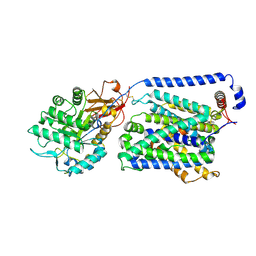 | | Overall structure of the y+LAT1-4F2hc bound with Arg | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARGININE, Amino acid transporter heavy chain SLC3A2, ... | | Authors: | Yan, R.H, Dai, L, Zhang, Y.Y, Zhang, T. | | Deposit date: | 2024-03-06 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the substrate recognition and transport mechanism of the human y + LAT1-4F2hc transporter complex.
Sci Adv, 11, 2025
|
|
9JVP
 
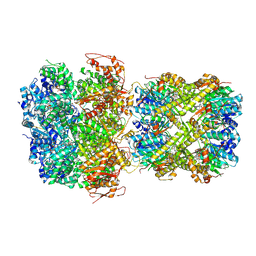 | | CryoEM structure of M. tuberculosis ClpC1P1P2 complex bound to bortezomib, conformation 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, ... | | Authors: | Zhou, B, Zhao, H, Gao, Y, Chen, X, Zhang, T, He, J, Xiong, X. | | Deposit date: | 2024-10-09 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Activation mechanism of caseinolytic chaperone-protease system in Mycobacterium tuberculosis by the anti-cancer drug bortezomib
To Be Published
|
|
9JVZ
 
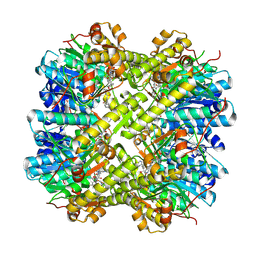 | | CryoEM structure of M. tuberculosis ClpP1P2 bound to bortezomib | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE | | Authors: | Zhou, B, Zhao, H, Gao, Y, Chen, W, Zhang, T, He, J, Xiong, X. | | Deposit date: | 2024-10-09 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Activation mechanism of caseinolytic chaperone-protease system in Mycobacterium tuberculosis by the anti-cancer drug bortezomib
To Be Published
|
|
9KY5
 
 | | Structure of y+LAT2 bound with Arg | | Descriptor: | ARGININE, Amino acid transporter heavy chain SLC3A2, Y+L amino acid transporter 2 | | Authors: | Yan, R.H, Dai, L, Zhang, T. | | Deposit date: | 2024-12-08 | | Release date: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of y+LAT2 bound with Arg
To Be Published
|
|
7VKR
 
 | |
7VKQ
 
 | |
7X9E
 
 | | Crystal structure of the 76E1 Fab in complex with a SARS-CoV-2 spike peptide | | Descriptor: | 76E1 Fab Heavy Chain, 76E1 Fab Light Chain, Spike peptide | | Authors: | Chen, X, Zhang, T, Ding, J, Sun, X, Sun, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Neutralization mechanism of a human antibody with pan-coronavirus reactivity including SARS-CoV-2.
Nat Microbiol, 7, 2022
|
|
7BTA
 
 | | Crystal structure of Rheb D60K mutant bound to GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Zhang, C, Zhang, T, Ding, J. | | Deposit date: | 2020-03-31 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis for the functions of dominantly active Y35N and inactive D60K Rheb mutants in mTORC1 signaling.
J Mol Cell Biol, 12, 2020
|
|
7BTC
 
 | | Crystal structure of Rheb Y35N mutant bound to GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Zhang, C, Zhang, T, Ding, J. | | Deposit date: | 2020-04-01 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Molecular basis for the functions of dominantly active Y35N and inactive D60K Rheb mutants in mTORC1 signaling.
J Mol Cell Biol, 12, 2020
|
|
8XYJ
 
 | | Structure of y+LAT1 bound with Lys | | Descriptor: | LYSINE, Y+L amino acid transporter 1 | | Authors: | Yan, R.H, Dai, L, Zhang, T. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural basis for the substrate recognition and transport mechanism of the human y + LAT1-4F2hc transporter complex.
Sci Adv, 11, 2025
|
|
