1U5R
 
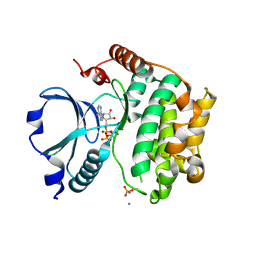 | | Crystal Structure of the TAO2 Kinase Domain: Activation and Specifity of a Ste20p MAP3K | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Zhou, T, Raman, M, Gao, Y, Earnest, S, Chen, Z, Machius, M, Cobb, M.H, Goldsmith, E.J. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the TAO2 Kinase Domain; Activation and Specificity of a Ste20p MAP3K.
Structure, 12, 2004
|
|
1W15
 
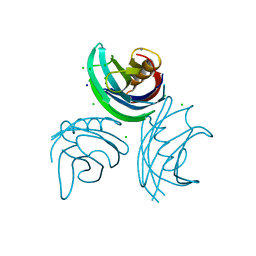 | | rat synaptotagmin 4 C2B domain in the presence of calcium | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Dai, H, Shin, O.-H, Machius, M, Tomchick, D.R, Sudhof, T.C, Rizo, J. | | Deposit date: | 2004-06-16 | | Release date: | 2004-08-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Basis for the Evolutionary Inactivation of Ca2+ Binding to Synaptotagmin 4
Nat.Struct.Mol.Biol., 11, 2004
|
|
1W16
 
 | | rat synaptotagmin 4 C2B domain in the absence of calcium | | Descriptor: | CHLORIDE ION, SODIUM ION, SYNAPTOTAGMIN IV | | Authors: | Dai, H, Shin, O.-H, Machius, M, Tomchick, D.R, Sudhof, T.C, Rizo, J. | | Deposit date: | 2004-06-16 | | Release date: | 2004-08-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Evolutionary Inactivation of Ca2+ Binding to Synaptotagmin 4
Nat.Struct.Mol.Biol., 11, 2004
|
|
1XF9
 
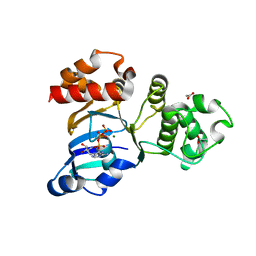 | | Structure of NBD1 from murine CFTR- F508S mutant | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, ... | | Authors: | Thibodeau, P.H, Brautigam, C.A, Machius, M, Thomas, P.J. | | Deposit date: | 2004-09-14 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Side chain and backbone contributions of Phe508 to CFTR folding.
Nat.Struct.Mol.Biol., 12, 2005
|
|
7UCC
 
 | | Transcription factor FosB/JunD bZIP domain in the reduced form | | Descriptor: | CHLORIDE ION, ETHANOL, Protein fosB, ... | | Authors: | Kumar, A, Machius, M.C, Rudenko, G. | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Chemically targeting the redox switch in AP1 transcription factor Delta FOSB.
Nucleic Acids Res., 50, 2022
|
|
7UCD
 
 | | Transcription factor FosB/JunD bZIP domain covalently modified with the cysteine-targeting alpha-haloketone compound Z2159931480 | | Descriptor: | 7-acetyl-4-methoxy-1-benzofuran-3(2H)-one, CHLORIDE ION, Protein fosB, ... | | Authors: | Kumar, A, Machius, M.C, Rudenko, G. | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Chemically targeting the redox switch in AP1 transcription factor Delta FOSB.
Nucleic Acids Res., 50, 2022
|
|
3BEN
 
 | |
5VPC
 
 | | Transcription factor FosB/JunD bZIP domain in its oxidized form, type-II crystal | | Descriptor: | CHLORIDE ION, Protein fosB, SODIUM ION, ... | | Authors: | Yin, Z, Machius, M.C, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
1HVX
 
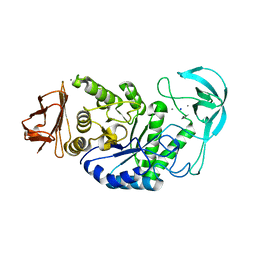 | | BACILLUS STEAROTHERMOPHILUS ALPHA-AMYLASE | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Suvd, D, Fujimoto, Z, Takase, K, Matsumura, M, Mizuno, H. | | Deposit date: | 2001-01-08 | | Release date: | 2001-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Bacillus stearothermophilus alpha-amylase: possible factors determining the thermostability.
J.Biochem., 129, 2001
|
|
4R8Q
 
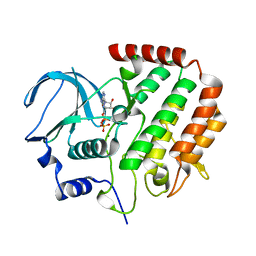 | |
3G5A
 
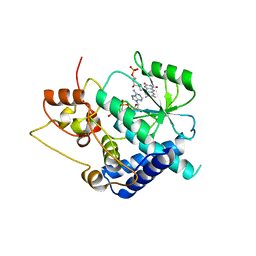 | |
6MEP
 
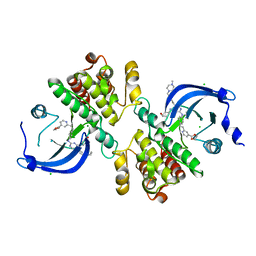 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC3437 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Tyrosine-protein kinase Mer, ... | | Authors: | Da, C, Zhang, D, Stashko, M.A, Cheng, A, Hunter, D, Norris-Drouin, J, Graves, L, Machius, M, Miley, M.J, DeRyckere, D, Earp, H.S, Graham, D.K, Frye, S.V, Wang, X, Kireev, D. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Data-Driven Construction of Antitumor Agents with Controlled Polypharmacology.
J.Am.Chem.Soc., 141, 2019
|
|
5C7I
 
 | |
4OQQ
 
 | |
3OBV
 
 | | Autoinhibited Formin mDia1 Structure | | Descriptor: | Protein diaphanous homolog 1, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Tomchick, D.R, Rosen, M.K, Otomo, T. | | Deposit date: | 2010-08-09 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the Formin mDia1 in autoinhibited conformation.
Plos One, 5, 2010
|
|
3BXH
 
 | |
3BXF
 
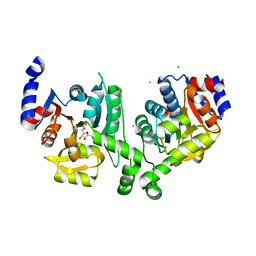 | | Crystal structure of effector binding domain of central glycolytic gene regulator (CggR) from Bacillus subtilis in complex with effector fructose-1,6-bisphosphate | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, 1,6-di-O-phosphono-beta-D-fructofuranose, CHLORIDE ION, ... | | Authors: | Rezacova, P, Otwinowski, Z. | | Deposit date: | 2008-01-13 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the effector-binding domain of repressor Central glycolytic gene Regulator from Bacillus subtilis reveal ligand-induced structural changes upon binding of several glycolytic intermediates.
Mol.Microbiol., 69, 2008
|
|
3BXG
 
 | |
3BXE
 
 | |
3GMH
 
 | | Crystal Structure of the Mad2 Dimer | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2A, SULFATE ION | | Authors: | Ozkan, E, Luo, X, Machius, M, Yu, H, Deisenhofer, J. | | Deposit date: | 2009-03-13 | | Release date: | 2010-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structure of an intermediate conformer of the spindle checkpoint protein Mad2.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2QYF
 
 | |
2OKG
 
 | | Structure of effector binding domain of central glycolytic gene regulator (CggR) from B. subtilis | | Descriptor: | CHLORIDE ION, Central glycolytic gene regulator, GLYCERALDEHYDE-3-PHOSPHATE | | Authors: | Rezacova, P, Moy, S.F, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-16 | | Release date: | 2007-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of the effector-binding domain of repressor Central glycolytic gene Regulator from Bacillus subtilis reveal ligand-induced structural changes upon binding of several glycolytic intermediates.
Mol.Microbiol., 69, 2008
|
|
4OQP
 
 | |
3EDV
 
 | |
3G59
 
 | |
