2H9W
 
 | |
6ENM
 
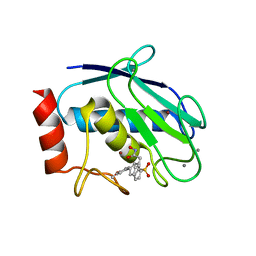 | | Crystal structure of MMP12 in complex with hydroxamate inhibitor LP168. | | Descriptor: | 2-[2-[4-(4-methoxyphenyl)phenyl]sulfonylphenyl]-~{N}-oxidanyl-ethanamide, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Vera, L, Nuti, E, Rossello, A, Stura, E.A. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Development of Thioaryl-Based Matrix Metalloproteinase-12 Inhibitors with Alternative Zinc-Binding Groups: Synthesis, Potentiometric, NMR, and Crystallographic Studies.
J. Med. Chem., 61, 2018
|
|
4RMS
 
 | |
2X89
 
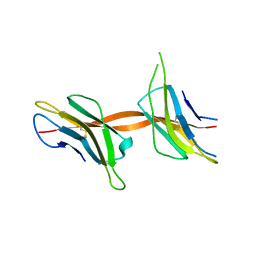 | | Structure of the Beta2_microglobulin involved in amyloidogenesis | | Descriptor: | ANTIBODY, BETA-2-MICROGLOBULIN | | Authors: | Domanska, K, Srinivasan, V, Vanderhaegen, S, Pardon, E, Marquez, J.A, Bellotti, V, Wyns, L, Steyaert, J. | | Deposit date: | 2010-03-07 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Atomic Structure of a Nanobody-Trapped Domain-Swapped Dimer of an Amyloidogenic {Beta}2-Microglobulin Variant.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4RMT
 
 | | Crystal structure of the D98N Beta-2 Microglobulin mutant | | Descriptor: | ACETATE ION, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | de Rosa, M, Bolognesi, M, Ricagno, S. | | Deposit date: | 2014-10-22 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.242 Å) | | Cite: | Decoding the Structural Bases of D76N 2-Microglobulin High Amyloidogenicity through Crystallography and Asn-Scan Mutagenesis.
Plos One, 10, 2015
|
|
4RMR
 
 | |
4RMQ
 
 | |
5DO6
 
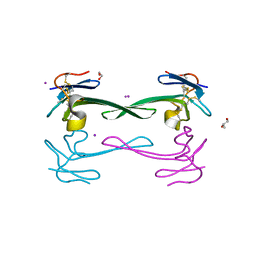 | | Crystal structure of Dendroaspis polylepis venom mambalgin-1 T23A mutant | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, Mambalgin-1, ... | | Authors: | Stura, E.A, Tepshi, L, Kessler, P, Gilles, M, Servent, D. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-30 | | Last modified: | 2017-01-25 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Mambalgin-1 Pain-relieving Peptide, Stepwise Solid-phase Synthesis, Crystal Structure, and Functional Domain for Acid-sensing Ion Channel 1a Inhibition.
J.Biol.Chem., 291, 2016
|
|
1RGJ
 
 | | NMR STRUCTURE OF THE COMPLEX BETWEEN ALPHA-BUNGAROTOXIN AND MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR WITH ENHANCED ACTIVITY | | Descriptor: | MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR, long neurotoxin 1 | | Authors: | Bernini, A, Spiga, O, Ciutti, A, Scarselli, M, Bracci, L, Lozzi, L, Lelli, B, Neri, P, Niccolai, N. | | Deposit date: | 2003-11-12 | | Release date: | 2003-11-25 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR and MD studies on the interaction between ligand peptides and alpha-bungarotoxin.
J.Mol.Biol., 339, 2004
|
|
7UBF
 
 | |
7UBD
 
 | |
7UBI
 
 | |
7UZL
 
 | |
7UBC
 
 | |
7UBE
 
 | |
7UBG
 
 | |
7UBH
 
 | |
8CTO
 
 | |
8CUN
 
 | |
8CWA
 
 | |
5A5Z
 
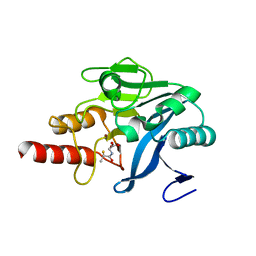 | | Approved Drugs Containing Thiols as Inhibitors of Metallo-beta- lactamases: Strategy To Combat Multidrug-Resistant Bacteria | | Descriptor: | BETA-LACTAMASE NDM-1, TIOPRONIN, ZINC ION | | Authors: | Klingler, F.M, Wichelhaus, T.A, Frank, D, Cuesta-Bernal, J, El-Delik, J, Mueller, H.F, Sjuts, H, Goettig, S, Koenigs, A, Pogoryelov, D, Proschak, E. | | Deposit date: | 2015-06-23 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Approved Drugs Containing Thiols as Inhibitors of Metallo-beta-lactamases: Strategy To Combat Multidrug-Resistant Bacteria.
J. Med. Chem., 58, 2015
|
|
4MRB
 
 | | Wild Type Human Transthyretin pH 7.5 | | Descriptor: | CALCIUM ION, Transthyretin | | Authors: | Chen, W.J, Wood, S.P. | | Deposit date: | 2013-09-17 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Proteolytic cleavage of Ser52Pro variant transthyretin triggers its amyloid fibrillogenesis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MRC
 
 | | Human Transthyretin Ser52Pro Mutant | | Descriptor: | CALCIUM ION, Transthyretin | | Authors: | Chen, W.J, Wood, S.P. | | Deposit date: | 2013-09-17 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Proteolytic cleavage of Ser52Pro variant transthyretin triggers its amyloid fibrillogenesis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6WHO
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, SODIUM ION, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WHN
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, DI(HYDROXYETHYL)ETHER, Histone deacetylase 2, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
