2AB6
 
 | |
6OI3
 
 | | Crystal structure of human WDR5 in complex with monomethyl H3R2 peptide | | Descriptor: | GLYCEROL, Monomethyl H3R2 peptide, SULFATE ION, ... | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
6OI1
 
 | | Crystal structure of human WDR5 in complex with monomethyl L-arginine | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
6OFZ
 
 | | Crystal structure of human WDR5 | | Descriptor: | WD repeat-containing protein 5 | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-01 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
6OI2
 
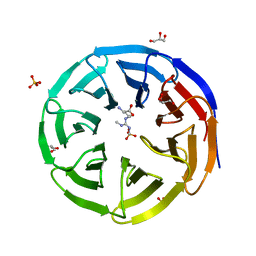 | | Crystal structure of human WDR5 in complex with symmetric dimethyl-L-arginine | | Descriptor: | GLYCEROL, N3, N4-DIMETHYLARGININE, ... | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
1AOA
 
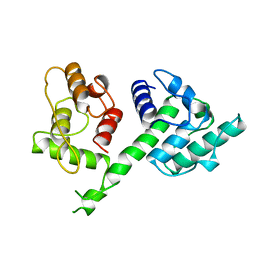 | | N-TERMINAL ACTIN-CROSSLINKING DOMAIN FROM HUMAN FIMBRIN | | Descriptor: | T-FIMBRIN | | Authors: | Goldsmith, S.C, Pokala, N, Shen, W, Fedorov, A.A, Matsudaira, P, Almo, S.C. | | Deposit date: | 1997-06-30 | | Release date: | 1997-12-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of an actin-crosslinking domain from human fimbrin.
Nat.Struct.Biol., 4, 1997
|
|
1B8N
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-1-(S)-(9-DEAZAGUANIN-9-YL)-D-RIBITOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Fedorov, A.A, Kicska, G.A, Fedorov, E.V, Strokopytov, B.V, Tyler, P.C, Furneaux, R.H, Schramm, V.L, Almo, S.C. | | Deposit date: | 1999-02-02 | | Release date: | 1999-02-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic dissection of the hydrogen bond network for transition-state analogue binding to purine nucleoside phosphorylase
Biochemistry, 41, 2002
|
|
1B8O
 
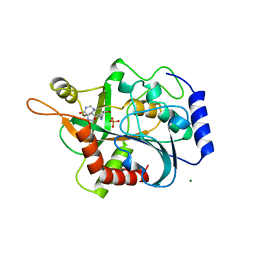 | | PURINE NUCLEOSIDE PHOSPHORYLASE | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Fedorov, A.A, Kicska, G.A, Fedorov, E.V, Shi, W, Tyler, P.C, Furneaux, R.H, Schramm, V.L, Almo, S.C. | | Deposit date: | 1999-02-02 | | Release date: | 1999-02-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Transition state structure of purine nucleoside phosphorylase and principles of atomic motion in enzymatic catalysis.
Biochemistry, 40, 2001
|
|
5DMH
 
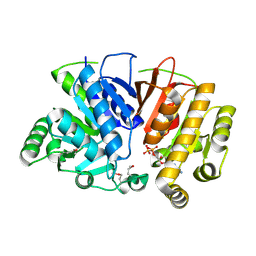 | | Crystal structure of a domain of unknown function (DUF1537) from Ralstonia eutropha H16 (H16_A1561), Target EFI-511666, complex with ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Uncharacterized protein conserved in bacteria | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-09-08 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a domain of unknown function (DUF1537) from Ralstonia eutropha H16 (H16_A1561), Target EFI-511666, complex with ADP.
To be published
|
|
4X24
 
 | | Crystal structure of Vibrio cholerae 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with methylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, TRIETHYLENE GLYCOL | | Authors: | Cameron, S.A, Thomas, K, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-11-25 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active site and remote contributions to catalysis in methylthioadenosine nucleosidases.
Biochemistry, 54, 2015
|
|
5TNV
 
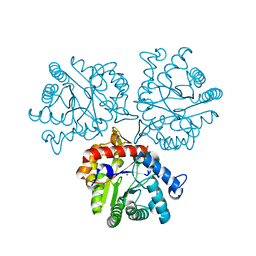 | | Crystal Structure of a Xylose isomerase-like TIM barrel Protein from Mycobacterium smegmatis in Complex with Magnesium | | Descriptor: | AP endonuclease, family protein 2, MAGNESIUM ION | | Authors: | Cook, W.J, Fedorov, A.A, Fedorov, E.V, Huang, H, Bonanno, J.B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2016-10-14 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystal Structure of a Xylose isomerase-like TIM barrel Protein from Mycobacterium smegmatis in Complex with Magnesium
To Be Published
|
|
5TVZ
 
 | | Solution NMR structure of Saccharomyces cerevisiae Pom152 Ig-like repeat, residues 718-820 | | Descriptor: | Nucleoporin POM152 | | Authors: | Dutta, K, Sampathkumar, P, Cowburn, D, Almo, S.C, Rout, M.P, Fernandez-Martinez, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex.
Structure, 25, 2017
|
|
5TX7
 
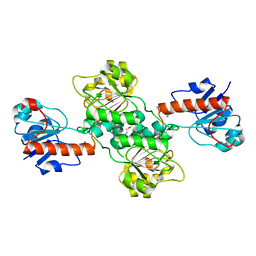 | | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Desulfovibrio vulgaris | | Descriptor: | D-isomer specific 2-hydroxyacid dehydrogenase family protein, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL | | Authors: | Czub, M.P, Shabalin, I.G, Gasiorowska, O.A, Handing, K.B, Kutner, J, Cymborowski, M.T, Hennig, P.M, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-11-15 | | Release date: | 2016-12-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Desulfovibrio vulgaris
to be published
|
|
5TRD
 
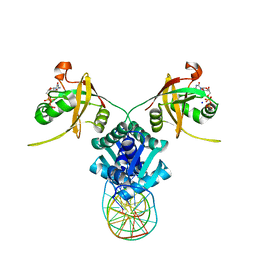 | | Structure of RbkR (Riboflavin Kinase) from Thermoplasma acidophilum determined in complex with CTP and its cognate DNA operator | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*TP*TP*AP*CP*TP*AP*AP*TP*TP*CP*AP*CP*GP*AP*GP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*AP*CP*TP*CP*GP*TP*GP*AP*AP*TP*TP*AP*GP*TP*AP*A)-3'), ... | | Authors: | Vetting, M.W, Rodionova, I.A, Li, X, Osterman, A.L, Rodionov, D.A, Almo, S.C. | | Deposit date: | 2016-10-26 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of RbkR (Riboflavin Kinase) from Thermoplasma acidophilum determined in complex with CTP and its cognate DNA operator
To be published
|
|
8SFZ
 
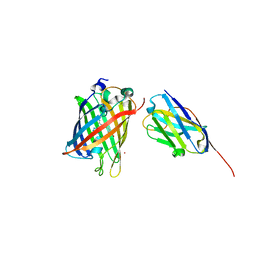 | | High Affinity nanobodies against GFP | | Descriptor: | Green fluorescent protein, LaG35, POTASSIUM ION, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unique mechanisms to increase structural stability and enhance antigen binding in nanobodies.
Structure, 33, 2025
|
|
8SFS
 
 | | High Affinity nanobodies against GFP | | Descriptor: | AMMONIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonnano, J.B, Almo, S.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Unique mechanisms to increase structural stability and enhance antigen binding in nanobodies.
Structure, 33, 2025
|
|
8SFV
 
 | | High affinity nanobodies to GFP | | Descriptor: | GLYCEROL, Green fluorescent protein, LaG19, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Unique mechanisms to increase structural stability and enhance antigen binding in nanobodies.
Structure, 33, 2025
|
|
8SLC
 
 | | High Affinity nanobodies against GFP | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2023-04-21 | | Release date: | 2024-05-22 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Unique mechanisms to increase structural stability and enhance antigen binding in nanobodies.
Structure, 33, 2025
|
|
8SFX
 
 | | High Affinity nanobodies against GFP | | Descriptor: | D-MALATE, GLYCEROL, Green fluorescent protein, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unique mechanisms to increase structural stability and enhance antigen binding in nanobodies.
Structure, 33, 2025
|
|
8SG3
 
 | | High Affinity nanobodies against GFP | | Descriptor: | Green fluorescent protein, LaG41 | | Authors: | Ketaren, N.E, Rout, M.P, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Unique mechanisms to increase structural stability and enhance antigen binding in nanobodies.
Structure, 33, 2025
|
|
5DM3
 
 | | Crystal Structure of Glutamine Synthetase from Chromohalobacter salexigens DSM 3043(Csal_0679, TARGET EFI-550015) with bound ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, L-glutamine synthetase | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-09-07 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Glutamine Synthetase from Chromohalobacter salexigens DSM 3043(Csal_0679, TARGET EFI-550015) with bound ADP
To be published
|
|
5E5M
 
 | | Crystal structure of mouse CTLA-4 in complex with nanobody | | Descriptor: | CTLA-4 nanobody, Cytotoxic T-lymphocyte protein 4, GLYCEROL | | Authors: | Fedorov, A.A, Fedorov, E.V, Samanta, D, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2015-10-08 | | Release date: | 2016-10-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Crystal structure of mouse CTLA-4 in complex with nanobody
To Be Published
|
|
5E83
 
 | | CRYSTAL STRUCTURE OF CARBONMONOXY HEMOGLOBIN S (LIGANDED SICKLE CELL HEMOGLOBIN) COMPLEXED WITH GBT440, CO-CRYSTALLIZATION EXPERIMENT | | Descriptor: | 2-methyl-3-({2-[1-(propan-2-yl)-1H-pyrazol-5-yl]pyridin-3-yl}methoxy)phenol, CARBON MONOXIDE, GLYCEROL, ... | | Authors: | Patskovska, L, Patskovsky, Y, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2015-10-13 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | GBT440 increases haemoglobin oxygen affinity, reduces sickling and prolongs RBC half-life in a murine model of sickle cell disease.
Br.J.Haematol., 175, 2016
|
|
5E56
 
 | | Crystal structure of mouse CTLA-4 | | Descriptor: | Cytotoxic T-lymphocyte protein 4, SODIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, SAMANTA, D, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2015-10-07 | | Release date: | 2015-10-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Crystal structure of mouse CTLA-4
To Be Published
|
|
5E03
 
 | | Crystal structure of mouse CTLA-4 nanobody | | Descriptor: | CTLA-4 nanobody, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Samanta, D, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.685 Å) | | Cite: | Crystal structure of mouse CTLA-4 nanobody
To Be Published
|
|
