9ITU
 
 | | Chloroflexus aurantiacus ADP-bound ATP synthase, state 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITS
 
 | | Chloroflexus aurantiacus ADP-bound ATP synthase, state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITR
 
 | | Chloroflexus aurantiacus ATP synthase, state 3, focused refinement of FO and peripheral stalk | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9IU0
 
 | | Chloroflexus aurantiacus ADP-bound ATP synthase, state 3, focused refinement of FO and peripheral stalk | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (5.18 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITL
 
 | | Chloroflexus aurantiacus ATP synthase, state 3 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITJ
 
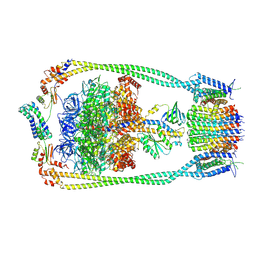 | | Chloroflexus aurantiacus ATP synthase, state 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITK
 
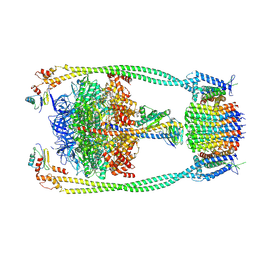 | | Chloroflexus aurantiacus ATP synthase, state 2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITM
 
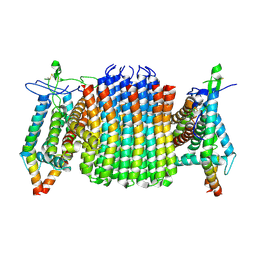 | | Chloroflexus aurantiacus ATP synthase, state 1, focused refinement of FO | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITV
 
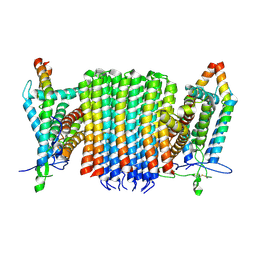 | | Chloroflexus aurantiacus ADP-bound ATP synthase, state 1, focused refinement of FO | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITQ
 
 | | Chloroflexus aurantiacus ATP synthase, state 3, focused refinement of FO | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITP
 
 | | Chloroflexus aurantiacus ATP synthase, state 2, focused refinement of FO and peripheral stalk | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITZ
 
 | | Chloroflexus aurantiacus ADP-bound ATP synthase, state 3, focused refinement of FO | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITY
 
 | | Chloroflexus aurantiacus ADP-bound ATP synthase, state 2, focused refinement of FO and peripheral stalk | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (4.95 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
6K5L
 
 | | The crystal structure of isocitrate dehydrogenase kinase/phosphatase wtih two Mn2+ from E. coli | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Isocitrate dehydrogenase kinase/phosphatase, ... | | Authors: | Zhang, X, Lei, Z, Zheng, J, Jia, Z. | | Deposit date: | 2019-05-29 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Characterization of metal binding of bifunctional kinase/phosphatase AceK and implication in activity modulation.
Sci Rep, 9, 2019
|
|
6J6A
 
 | |
9BYO
 
 | | Cryo-EM structure of glucagon-like peptide-1 receptor (GLP-1R)-Gs complex with Exendin-asp3 | | Descriptor: | Exendin-3, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Johnson, R, Belousoff, M.J, Danev, R, Sexton, P.M, Wootten, D. | | Deposit date: | 2024-05-24 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | Structural insights in the activation of GLP-1R by biased ligands
To Be Published
|
|
9C0K
 
 | | Cryo-EM structure of glucagon-like peptide-1 receptor (GLP-1R)-Gs complex with Exendin-phe1 | | Descriptor: | Exendin-4, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Johnson, R, Belousoff, M.J, Danev, R, Sexton, P.M, Wootten, D. | | Deposit date: | 2024-05-25 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural insights into the activation of GLP-1R by biased ligands
To Be Published
|
|
8H0Z
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-122 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
5GTX
 
 | | Crystal structure of mutated buckwheat glutaredoxin | | Descriptor: | buckwheat glutaredoxin | | Authors: | Zhang, X, Wang, W, Zhao, Y, Wang, Z, Wang, H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural insights into the binding of buckwheat glutaredoxin with GSH and regulation of its catalytic activity
J. Inorg. Biochem., 173, 2017
|
|
2GS6
 
 | | Crystal Structure of the active EGFR kinase domain in complex with an ATP analog-peptide conjugate | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, Peptide, ... | | Authors: | Zhang, X, Gureasko, J, Shen, K, Cole, P.A, Kuriyan, J. | | Deposit date: | 2006-04-25 | | Release date: | 2006-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An Allosteric Mechanism for Activation of the Kinase Domain of Epidermal Growth Factor Receptor
Cell(Cambridge,Mass.), 125, 2006
|
|
2GS7
 
 | | Crystal Structure of the inactive EGFR kinase domain in complex with AMP-PNP | | Descriptor: | Epidermal growth factor receptor, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Zhang, X, Gureasko, J, Shen, K, Cole, P.A, Kuriyan, J. | | Deposit date: | 2006-04-25 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor
Cell(Cambridge,Mass.), 125, 2006
|
|
2GS2
 
 | | Crystal Structure of the active EGFR kinase domain | | Descriptor: | Epidermal growth factor receptor | | Authors: | Zhang, X, Gureasko, J, Shen, K, Cole, P.A, Kuriyan, J. | | Deposit date: | 2006-04-25 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor.
Cell(Cambridge,Mass.), 125, 2006
|
|
8G05
 
 | | Cryo-EM structure of an orphan GPCR-Gi protein signaling complex | | Descriptor: | 6-(octylamino)pyrimidine-2,4(3H,5H)-dione, CHOLESTEROL, G-protein coupled receptor 84, ... | | Authors: | Zhang, X, Wang, Y.J, Li, X, Liu, G.B, Gong, W.M, Zhang, C. | | Deposit date: | 2023-01-31 | | Release date: | 2023-11-01 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Pro-phagocytic function and structural basis of GPR84 signaling.
Nat Commun, 14, 2023
|
|
5T2C
 
 | | CryoEM structure of the human ribosome at 3.6 Angstrom resolution | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Zhang, X, Lai, M, Zhou, Z.H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-01-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures and stabilization of kinetoplastid-specific split rRNAs revealed by comparing leishmanial and human ribosomes.
Nat Commun, 7, 2016
|
|
3F4M
 
 | | Crystal structure of TIPE2 | | Descriptor: | CHLORIDE ION, Tumor necrosis factor, alpha-induced protein 8-like protein 2 | | Authors: | Zhang, X, Wang, J, Shi, Y. | | Deposit date: | 2008-11-01 | | Release date: | 2009-03-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Crystal structure of TIPE2 provides insights into immune homeostasis
Nat.Struct.Mol.Biol., 16, 2009
|
|
