4KGC
 
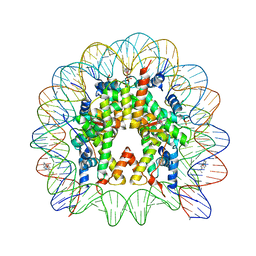 | | Nucleosome Core Particle Containing (ETA6-P-CYMENE)-(1, 2-ETHYLENEDIAMINE)-RUTHENIUM | | Descriptor: | (ethane-1,2-diamine-kappa~2~N,N')[(1,2,3,4,5,6-eta)-1-methyl-4-(propan-2-yl)cyclohexane-1,2,3,4,5,6-hexayl]ruthenium, DNA (145-mer), Histone H2A, ... | | Authors: | Adhireksan, Z, Davey, C.A. | | Deposit date: | 2013-04-29 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Ligand substitutions between ruthenium-cymene compounds can control protein versus DNA targeting and anticancer activity
Nat Commun, 5, 2014
|
|
4AMQ
 
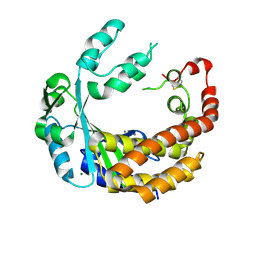 | |
1U79
 
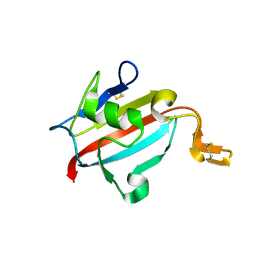 | | Crystal structure of AtFKBP13 | | Descriptor: | FKBP-type peptidyl-prolyl cis-trans isomerase 3 | | Authors: | Gopalan, G, Swaminathan, K. | | Deposit date: | 2004-08-03 | | Release date: | 2004-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis uncovers a role for redox in regulating FKBP13, an immunophilin of the chloroplast thylakoid lumen
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
4GJ2
 
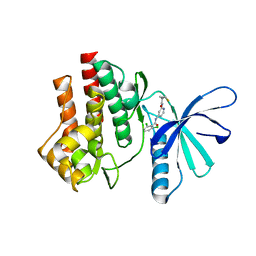 | | Tyk2 (JH1) in complex with 2,6-dichloro-N-[2-({[(1R,2R)-2-fluorocyclopropyl]carbonyl}amino)pyridin-4-yl]benzamide | | Descriptor: | 2,6-dichloro-N-[2-({[(1R,2R)-2-fluorocyclopropyl]carbonyl}amino)pyridin-4-yl]benzamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Ultsch, M.H. | | Deposit date: | 2012-08-09 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lead Optimization of a 4-Aminopyridine Benzamide Scaffold To Identify Potent, Selective, and Orally Bioavailable TYK2 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4NXO
 
 | | Crystal Structure of Insulin Degrading Enzyme in complex with BDM44768 | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Liang, W.G, Deprez, R, Deprez, B, Tang, W. | | Deposit date: | 2013-12-09 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice.
Nat Commun, 6, 2015
|
|
8OTM
 
 | | structure of InhA from mycobacterium tuberculosis in complex with N-((1-(3-hydroxy-4-phenoxybenzyl)-1H-1,2,3-triazol-4-yl)methyl)-2-oxo-2H-chromene-3-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 2-oxidanylidene-~{N}-[[1-[(3-oxidanyl-4-phenoxy-phenyl)methyl]-1,2,3-triazol-4-yl]methyl]chromene-3-carboxamide, ACETATE ION, ... | | Authors: | Chebaiki, M, Maveyraud, L, Tamhaev, R, Lherbet, C, Mourey, L. | | Deposit date: | 2023-04-21 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of new diaryl ether inhibitors against Mycobacterium tuberculosis targeting the minor portal of InhA.
Eur.J.Med.Chem., 259, 2023
|
|
8OTN
 
 | | structure of InhA from mycobacterium tuberculosis in complex with inhibitor 7-((1-(3-Hydroxy-4-phenoxybenzyl)-1H-1,2,3-triazol-4-yl)methoxy)-4-methyl-2H-chromen-2-one | | Descriptor: | 4-methyl-7-[[1-[(3-oxidanyl-4-phenoxy-phenyl)methyl]-1,2,3-triazol-4-yl]methoxy]chromen-2-one, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Chebaiki, M, Maveyraud, L, Tamhaev, R, Lherbet, C, Mourey, L. | | Deposit date: | 2023-04-21 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Discovery of new diaryl ether inhibitors against Mycobacterium tuberculosis targeting the minor portal of InhA.
Eur.J.Med.Chem., 259, 2023
|
|
6S05
 
 | |
6RZZ
 
 | |
3D11
 
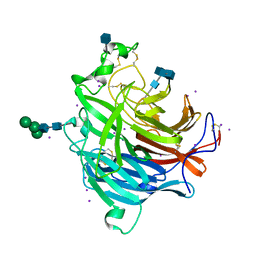 | | Crystal Structures of the Nipah G Attachment Glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-neuraminidase, ... | | Authors: | Xu, K, Rajashankar, K.R, Chan, Y.P, Himanen, P, Broder, C.C, Nikolov, D.B. | | Deposit date: | 2008-05-02 | | Release date: | 2008-08-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4I2Y
 
 | |
2BRE
 
 | | STRUCTURE OF A HSP90 INHIBITOR BOUND TO THE N-TERMINUS OF YEAST HSP90. | | Descriptor: | 4-{4-[4-(3-AMINOPROPOXY)PHENYL]-1H-PYRAZOL-5-YL}-6-CHLOROBENZENE-1,3-DIOL, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Roe, S.M, Pearl, L.H, Prodromou, C. | | Deposit date: | 2005-05-04 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The identification, synthesis, protein crystal structure and in vitro biochemical evaluation of a new 3,4-diarylpyrazole class of Hsp90 inhibitors.
Bioorg. Med. Chem. Lett., 15, 2005
|
|
6SUR
 
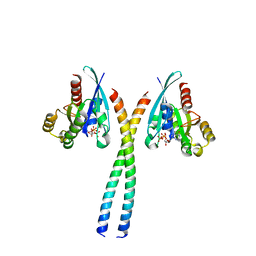 | | The Rab33B-Atg16L1 crystal structure | | Descriptor: | Autophagy-related protein 16-1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Metje-Sprink, J, Kuehnel, K. | | Deposit date: | 2019-09-16 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.467 Å) | | Cite: | Crystal structure of the Rab33B/Atg16L1 effector complex.
Sci Rep, 10, 2020
|
|
2W6O
 
 | | Crystal structure of Biotin carboxylase from E. coli in complex with 4-Amino-7,7-dimethyl-7,8-dihydro-quinazolinone fragment | | Descriptor: | 4-amino-7,7-dimethyl-7,8-dihydroquinazolin-5(6H)-one, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-12-18 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Antibacterial Biotin Carboxylase Inhibitors by Virtual Screening and Fragment-Based Approaches.
Acs Chem.Biol., 4, 2009
|
|
2BRC
 
 | | Structure of a Hsp90 Inhibitor bound to the N-terminus of Yeast Hsp90. | | Descriptor: | 4-[4-(2,3-DIHYDRO-1,4-BENZODIOXIN-6-YL)-3-METHYL-1H-PYRAZOL-5-YL]-6-ETHYLBENZENE-1,3-DIOL, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Roe, S.M, Pearl, L.H, Prodromou, C. | | Deposit date: | 2005-05-04 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The identification, synthesis, protein crystal structure and in vitro biochemical evaluation of a new 3,4-diarylpyrazole class of Hsp90 inhibitors.
Bioorg. Med. Chem. Lett., 15, 2005
|
|
8QNZ
 
 | | Crystal Structure of a Class D Carbapenemase Complexed with Hydrolyzed Imipenem | | Descriptor: | (2R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-(2-methanimidamidoethylsulfanyl)-2,3-dihydro-1H-pyrrole -5-carboxylic acid, 1-BUTANOL, BROMIDE ION, ... | | Authors: | Zhou, Q, He, Y, Jin, Y. | | Deposit date: | 2023-09-27 | | Release date: | 2023-11-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | An Ion-Pair Induced Intermediate Complex Captured in Class D Carbapenemase Reveals Chloride Ion as a Janus Effector Modulating Activity.
Acs Cent.Sci., 9, 2023
|
|
2W6Z
 
 | |
2W6Q
 
 | | Crystal structure of Biotin carboxylase from E. coli in complex with the triazine-2,4-diamine fragment | | Descriptor: | 6-(2-phenoxyethoxy)-1,3,5-triazine-2,4-diamine, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-12-18 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Antibacterial Biotin Carboxylase Inhibitors by Virtual Screening and Fragment-Based Approaches.
Acs Chem.Biol., 4, 2009
|
|
2W71
 
 | |
2W70
 
 | |
2W6P
 
 | |
2W6M
 
 | |
8R1C
 
 | | SD1-2 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SD1-2 fab heavy chain, SD1-2 fab light chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2023-11-01 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | The SARS-CoV-2 neutralizing antibody response to SD1 and its evasion by BA.2.86.
Nat Commun, 15, 2024
|
|
8R1D
 
 | | SD1-3 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SD1-3 Fab Heavy Chain, SD1-3 Fab Light Chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2023-11-01 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | The SARS-CoV-2 neutralizing antibody response to SD1 and its evasion by BA.2.86.
Nat Commun, 15, 2024
|
|
8QZR
 
 | | SARS-CoV-2 delta RBD complexed with BA.4/5-9 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BA.4/5-9 heavy chain, BA.4/5-9 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-10-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
