6SS4
 
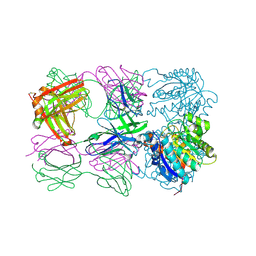 | | Structure of arginase-2 in complex with the inhibitory human antigen-binding fragment Fab C0021181 | | Descriptor: | Arginase-2, mitochondrial, Fab C0021181 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
2LU0
 
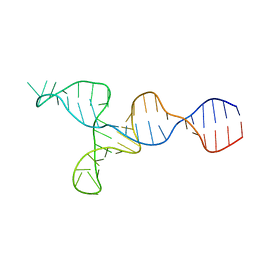 | | NMR solution structure of the kappa-zeta region of S.cerevisiae group II intron ai5(gamma) | | Descriptor: | RNA (49-MER) | | Authors: | Donghi, D, Pechlaner, M, Finazzo, C, Knobloch, B, Sigel, R.K.O. | | Deposit date: | 2012-06-05 | | Release date: | 2013-04-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structural stabilization of the kappa three-way junction by Mg(II) represents the first step in the folding of a group II intron.
Nucleic Acids Res., 41, 2013
|
|
7ZYM
 
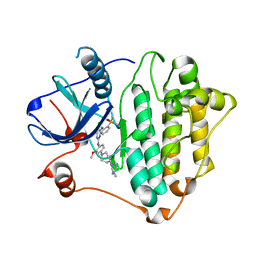 | | Crystal Structure of EGFR-T790M/C797S in Complex with Brigatinib | | Descriptor: | 5-chloro-N~4~-[2-(dimethylphosphoryl)phenyl]-N~2~-{2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl}pyrimidine-2,4-diamine, Epidermal growth factor receptor | | Authors: | Niggenaber, J, Kleinboelting, S, Mueller, M.P, Rauh, D. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Addressing the Osimertinib Resistance Mutation EGFR-L858R/C797S with Reversible Aminopyrimidines.
Acs Med.Chem.Lett., 14, 2023
|
|
3FCT
 
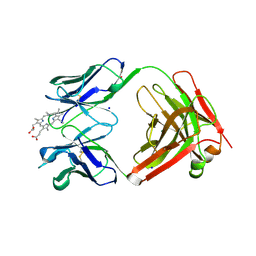 | | MATURE METAL CHELATASE CATALYTIC ANTIBODY WITH HAPTEN | | Descriptor: | CADMIUM ION, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Romesberg, F.E, Santarsiero, B.D, Barnes, D, Yin, J, Spiller, B, Schultz, P.G, Stevens, R.C. | | Deposit date: | 1999-06-13 | | Release date: | 1999-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and kinetic evidence for strain in biological catalysis.
Biochemistry, 37, 1998
|
|
7ZYN
 
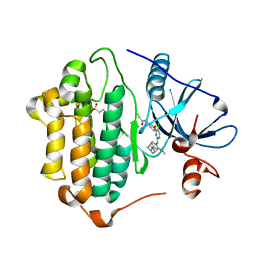 | | Crystal Structure of EGFR-T790M/C797S in Complex with WZ4002 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Epidermal growth factor receptor, N-{3-[(5-chloro-2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)oxy]phenyl}prop-2-enamide | | Authors: | Niggenaber, J, Kleinboelting, S, Mueller, M.P, Rauh, D. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Addressing the Osimertinib Resistance Mutation EGFR-L858R/C797S with Reversible Aminopyrimidines.
Acs Med.Chem.Lett., 14, 2023
|
|
7ZYP
 
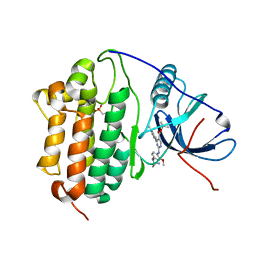 | | Crystal Structure of EGFR-T790M/C797S in Complex with Reversible Aminopyrimidine 9 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Epidermal growth factor receptor, propan-2-yl 2-[[4-(4-azanylpiperidin-1-yl)-2-methoxy-phenyl]amino]-4-(1-methylindol-3-yl)pyrimidine-5-carboxylate | | Authors: | Niggenaber, J, Kleinboelting, S, Mueller, M.P, Rauh, D. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Addressing the Osimertinib Resistance Mutation EGFR-L858R/C797S with Reversible Aminopyrimidines.
Acs Med.Chem.Lett., 14, 2023
|
|
3W0S
 
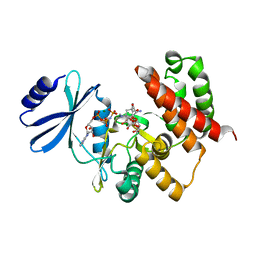 | | Crystal structure of aminoglycoside phosphotransferase APH(4)-Ia, ternary complex with AMP-PNP and hygromycin B | | Descriptor: | HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase, MAGNESIUM ION, ... | | Authors: | Iino, D, Takakura, Y, Fukano, K, Sasaki, Y, Hoshino, T, Ohsawa, K, Nakamura, A, Yajima, S. | | Deposit date: | 2012-11-02 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structures of the ternary complex of APH(4)-Ia/Hph with hygromycin B and an ATP analog using a thermostable mutant.
J.Struct.Biol., 183, 2013
|
|
3FFL
 
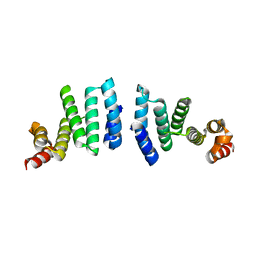 | |
7ZYQ
 
 | | Crystal Structure of EGFR-T790M/V948R in Complex with Reversible Aminopyrimidine 13 | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, SODIUM ION, ... | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Addressing the Osimertinib Resistance Mutation EGFR-L858R/C797S with Reversible Aminopyrimidines.
Acs Med.Chem.Lett., 14, 2023
|
|
3W0N
 
 | | Crystal structure of a thermostable mutant of aminoglycoside phosphotransferase APH(4)-Ia, ternary complex with AMP-PNP and hygromycin B | | Descriptor: | HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Iino, D, Takakura, Y, Fukano, K, Sasaki, Y, Hoshino, T, Ohsawa, K, Nakamura, A, Yajima, S. | | Deposit date: | 2012-11-02 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the ternary complex of APH(4)-Ia/Hph with hygromycin B and an ATP analog using a thermostable mutant.
J.Struct.Biol., 183, 2013
|
|
3W15
 
 | | Structure of peroxisomal targeting signal 2 (PTS2) of Saccharomyces cerevisiae 3-ketoacyl-CoA thiolase in complex with Pex7p and Pex21p | | Descriptor: | 3-ketoacyl-CoA thiolase, peroxisomal, Maltose-binding periplasmic protein, ... | | Authors: | Pan, D, Nakatsu, T, Kato, H. | | Deposit date: | 2012-11-06 | | Release date: | 2013-07-03 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of peroxisomal targeting signal-2 bound to its receptor complex Pex7p-Pex21p
Nat.Struct.Mol.Biol., 20, 2013
|
|
3FHW
 
 | | Crystal structure of the protein priB from Bordetella parapertussis. Northeast Structural Genomics Consortium target BpR162. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Primosomal replication protein n, SODIUM ION | | Authors: | Kuzin, A.P, Neely, H, Seetharaman, J, Forouhar, F, Wang, D, Mao, L, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-10 | | Release date: | 2008-12-30 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the protein priB from Bordetella parapertussis. Northeast Structural Genomics Consortium target BpR162.
To be Published
|
|
6SNZ
 
 | | Crystal structure of lamin A coil1b tetramer | | Descriptor: | GLYCEROL, Prelamin-A/C | | Authors: | Lilina, A.V, Chernyatina, A.A, Guzenko, D, Strelkov, S.V. | | Deposit date: | 2019-08-28 | | Release date: | 2019-10-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Lateral A11type tetramerization in lamins.
J.Struct.Biol., 209, 2020
|
|
2M0E
 
 | |
2MAB
 
 | |
3W0M
 
 | | Crystal structure of a thermostable mutant of aminoglycoside phosphotransferase APH(4)-Ia, apo form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Hygromycin-B 4-O-kinase | | Authors: | Iino, D, Takakura, Y, Fukano, K, Sasaki, Y, Hoshino, T, Ohsawa, K, Nakamura, A, Yajima, S. | | Deposit date: | 2012-11-02 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the ternary complex of APH(4)-Ia/Hph with hygromycin B and an ATP analog using a thermostable mutant.
J.Struct.Biol., 183, 2013
|
|
1DIU
 
 | | DIHYDROFOLATE REDUCTASE (E.C.1.5.1.3) COMPLEX WITH BRODIMOPRIM-4,6-DICARBOXYLATE | | Descriptor: | BRODIMOPRIM-4,6-DICARBOXYLATE, DIHYDROFOLATE REDUCTASE | | Authors: | Morgan, W.D, Birdsall, B, Polshakov, V.I, Sali, D, Kompis, I, Feeney, J. | | Deposit date: | 1995-08-01 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a brodimoprim analogue in its complex with Lactobacillus casei dihydrofolate reductase.
Biochemistry, 34, 1995
|
|
6SGU
 
 | | Cryo-EM structure of Escherichia coli AcrB and DARPin in Saposin A-nanodisc | | Descriptor: | DARPin, Multidrug efflux pump subunit AcrB | | Authors: | Szewczak-Harris, A, Du, D, Newman, C, Neuberger, A, Luisi, B.F. | | Deposit date: | 2019-08-05 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Interactions of a Bacterial RND Transporter with a Transmembrane Small Protein in a Lipid Environment.
Structure, 28, 2020
|
|
7ZNP
 
 | | Structure of AmedSP | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Sucrose phosphorylase | | Authors: | Fredslund, F, Teze, D, Welner, D.H. | | Deposit date: | 2022-04-21 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of AmedSP
To Be Published
|
|
6SPF
 
 | | Pseudomonas aeruginosa 70s ribosome from an aminoglycoside resistant clinical isolate | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-23 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3ISR
 
 | | The Crystal Structure of a Putative Cysteine Protease from Cytophaga hutchinsonii to 1.9A | | Descriptor: | Transglutaminase-like enzymes, putative cysteine protease | | Authors: | Stein, A.J, Bigelow, L, Trevino, D, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-27 | | Release date: | 2009-09-08 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of a Putative Cysteine Protease from Cytophaga hutchinsonii to 1.9A
To be Published
|
|
3ITA
 
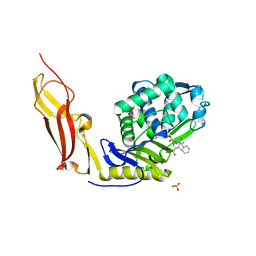 | | Crystal structure of Penicillin-Binding Protein 6 (PBP6) from E. coli in acyl-enzyme complex with ampicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, D-alanyl-D-alanine carboxypeptidase dacC, ... | | Authors: | Chen, Y, Zhang, W, Shi, Q, Hesek, D, Lee, M, Mobashery, S, Shoichet, B.K. | | Deposit date: | 2009-08-27 | | Release date: | 2009-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of penicillin-binding protein 6 from Escherichia coli.
J.Am.Chem.Soc., 131, 2009
|
|
6S8S
 
 | |
2M27
 
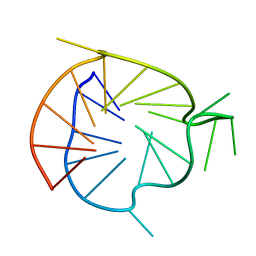 | | Major G-quadruplex structure formed in human VEGF promoter, a monomeric parallel-stranded quadruplex | | Descriptor: | DNA_(5'-D(*CP*GP*GP*GP*GP*CP*GP*GP*GP*CP*CP*TP*TP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3')_ | | Authors: | Agrawal, P, Hatzakis, E, Guo, K, Carver, M, Yang, D. | | Deposit date: | 2012-12-14 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: insights into loop interactions of the parallel G-quadruplexes.
Nucleic Acids Res., 41, 2013
|
|
2M3V
 
 | |
