4IP4
 
 | | Crystal structure of l-fuconate dehydratase from Silicibacter sp. tm1040 liganded with Mg | | Descriptor: | CARBON DIOXIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-01-09 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.128 Å) | | Cite: | Crystal structure of l-fuconate dehydratase from Silicibacter sp. tm1040 liganded with mg
To be Published
|
|
4JWT
 
 | |
4JXK
 
 | | Crystal Structure of Oxidoreductase ROP_24000 (Target EFI-506400) from Rhodococcus opacus B4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, oxidoreductase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-28 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Oxidoreductase Rop_24000 (Target EFI-506400) from Rhodococcus Opacus
To be Published
|
|
4JXY
 
 | | CRYSTAL STRUCTURE OF POLYPRENYL SYNTHASE PATL_3739 (TARGET EFI-509195) FROM Pseudoalteromonas atlantica | | Descriptor: | GLUTARIC ACID, PHOSPHATE ION, Trans-hexaprenyltranstransferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-28 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRYSTAL STRUCTURE OF ISOPRENOID SYNTHASE PATL_3739 FROM Pseudoalteromonas atlantica
To be Published
|
|
4J9W
 
 | | Crystal structure of the complex of a hydroxyproline epimerase (TARGET EFI-506499, PSEUDOMONAS FLUORESCENS PF-5) with the inhibitor pyrrole-2-carboxylate | | Descriptor: | GLYCEROL, PYRROLE-2-CARBOXYLATE, Proline racemase family protein, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-17 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the complex of a hydroxyproline epimerase (TARGET EFI-506499, PSEUDOMONAS FLUORESCENS PF-5) with the inhibitor pyrrole-2-carboxylate
To be Published
|
|
4JBD
 
 | | Crystal structure of Pput_1285, a putative hydroxyproline epimerase from Pseudomonas putida f1 (target EFI-506500), open form, space group I2, bound citrate | | Descriptor: | CITRIC ACID, Proline racemase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of pput_1285, a putative hydroxyproline epimerase from pseudomonas putida f1 (target efi-506500), open form, space group i2, bound citrate
To be Published
|
|
4JBB
 
 | | Crystal structure of Glutathione S-transferase A6TBY7(Target EFI-507184) from Klebsiella pneumoniae MGH 78578, GSH complex | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLUTATHIONE, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase A6Tby7 (Target Efi-507184) from Klebsiella Pneumoniae
To be Published
|
|
4JFC
 
 | | Crystal structure of a enoyl-CoA hydratase from Polaromonas sp. JS666 | | Descriptor: | Enoyl-CoA hydratase, GLYCEROL | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-28 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of a enoyl-CoA hydratase from Polaromonas sp. JS666
To be Published
|
|
4JLQ
 
 | | Crystal structure of human Karyopherin-beta2 bound to the PY-NLS of Saccharomyces cerevisiae NAB2 | | Descriptor: | Nuclear polyadenylated RNA-binding protein NAB2, Transportin-1 | | Authors: | Sampathkumar, P, Gizzi, A, Rout, M.P, Chook, Y.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of human Karyopherin beta 2 bound to the PY-NLS of Saccharomyces cerevisiae Nab2.
J.Struct.Funct.Genom., 14, 2013
|
|
4JD7
 
 | | Crystal structure of pput_1285, a putative hydroxyproline epimerase from Pseudomonas putida f1 (target EFI-506500), open form, space group P212121, bound sulfate | | Descriptor: | Proline racemase, SULFATE ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-24 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of pput_1285, a putative hydroxyproline epimerase from Pseudomonas putida f1 (target EFI-506500), open form, space group P212121, bound sulfate
To be Published
|
|
4JED
 
 | | Crystal structure of glutathione s-transferase mrad2831_1084 (target efi-507060) from methylobacterium radiotolerans jcm 2831, complex with glutathione sulfonate | | Descriptor: | GLUTATHIONE S-TRANSFERASE, GLUTATHIONE SULFONIC ACID, GLYCEROL | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-26 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of glutathione S-transferase (TARGET EFI-507060) from Methylobacterium radiotolerans
To be Published
|
|
4HCD
 
 | | Crystal structure of D-glucarate dehydratase from agrobacterium tumefaciens complexed with magnesium | | Descriptor: | CHLORIDE ION, Isomerase/lactonizing enzyme, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Millikin, C, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-09-29 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of D-glucarate dehydratase from agrobacterium tumefaciens complexed with magnesium
To be Published
|
|
4H83
 
 | | Crystal structure of Mandelate racemase/muconate lactonizing enzyme (EFI target:502127) | | Descriptor: | BICARBONATE ION, GLYCEROL, Mandelate racemase/muconate lactonizing enzyme, ... | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Crystal structure of Mandelate racemase/muconate lactonizing enzyme
To be Published
|
|
4H16
 
 | |
4H2H
 
 | | Crystal structure of an enolase (mandalate racemase subgroup, target EFI-502101) from Pelagibaca bermudensis htcc2601, with bound mg and l-4-hydroxyproline betaine (betonicine) | | Descriptor: | (2S,4R)-4-hydroxy-1,1-dimethylpyrrolidinium-2-carboxylate, (4S)-2-METHYL-2,4-PENTANEDIOL, IODIDE ION, ... | | Authors: | Vetting, M.W, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-09-12 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of new enzymes and metabolic pathways by using structure and genome context.
Nature, 502, 2013
|
|
4HB9
 
 | |
4HAD
 
 | | Crystal structure of probable oxidoreductase protein from Rhizobium etli CFN 42 | | Descriptor: | Probable oxidoreductase protein, SODIUM ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-09-26 | | Release date: | 2012-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of probable oxidoreductase protein from Rhizobium etli CFN 42
To be Published
|
|
4GRA
 
 | | Crystal structure of SULT1A1 bound with PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase 1A1 | | Authors: | Kim, J, Cook, I, Wang, T, Falany, C.N, Leyh, T.S, Almo, S.C. | | Deposit date: | 2012-08-24 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The gate that governs sulfotransferase selectivity.
Biochemistry, 52, 2013
|
|
4GVX
 
 | | Crystal structure of a short chain dehydrogenase homolog (target EFI-505321) from burkholderia multivorans, with bound NADP and L-fucose | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier protein] reductase, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Hobbs, M.E, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Crystal structure of a short chain dehydrogenase homolog (target EFI-505321) from burkholderia multivorans, with bound NADP and L-fucose
To be Published
|
|
4HC8
 
 | |
4HCL
 
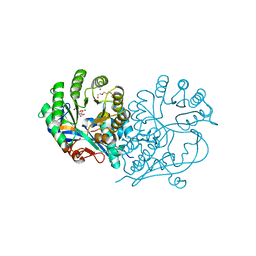 | | CRYSTAL STRUCTURE OF D-GLUCARATE DEHYDRATASE FROM AGROBACTERIUM TUMEFACIENS complexed with magnesium and L-Lyxarohydroxamate | | Descriptor: | (2R,3S,4R)-2,3,4-TRIHYDROXY-5-(HYDROXYAMINO)-5-OXOPENTANOIC ACID, Isomerase/lactonizing enzyme, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-09-30 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF D-GLUCARATE DEHYDRATASE FROM AGROBACTERIUM TUMEFACIENS complexed with magnesium and L-Lyxarohydroxamate
To be Published
|
|
4HL7
 
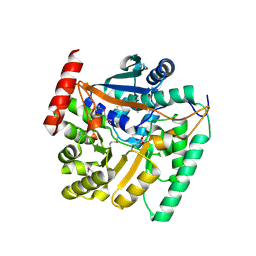 | | Crystal structure of nicotinate phosphoribosyltransferase (target NYSGR-026035) from Vibrio cholerae | | Descriptor: | Nicotinate phosphoribosyltransferase, SULFATE ION | | Authors: | Mulichak, A.M, Sauder, J.M, Keefe, L.J, Burley, S.K, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-10-16 | | Release date: | 2012-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of nicotinate phosphoribosyltransferase (target NYSGR-026035) from Vibrio cholerae
To be Published
|
|
4JXU
 
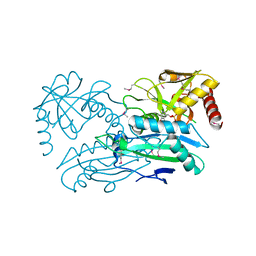 | | Structure of aminotransferase ilvE2 from Sinorhizobium meliloti complexed with PLP | | Descriptor: | Putative aminotransferase | | Authors: | Cooper, D.R, Cymborowski, M.T, Majorek, K.A, Niedzialkowska, E, Porebski, P.J, Stead, M, Hammonds, J, Seidel, R, Ahmed, M, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-28 | | Release date: | 2013-05-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of aminotransferase ilvE2 from Sinorhizobium meliloti complexed with PLP
To be Published
|
|
4JYX
 
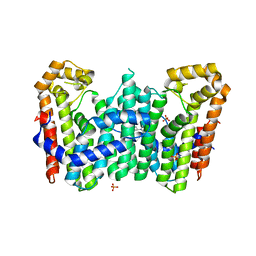 | | Crystal structure of polyprenyl synthase PATL_3739 (TARGET EFI-509195) FROM PSEUDOALTEROMONAS ATLANTICA, COMPLEX WITH INORGANIC PHOSPHATE AND AN UNKNOWN LIGAND | | Descriptor: | PHOSPHATE ION, Trans-hexaprenyltranstransferase, UNKNOWN LIGAND | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-04-01 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Isoprenoid Synthase Patl_3739 from Pseudoalteromonas Atlantica
To be Published
|
|
4K29
 
 | | Crystal structure of an enoyl-CoA hydratase/isomerase from Xanthobacter autotrophicus Py2 | | Descriptor: | Enoyl-CoA hydratase/isomerase, GLYCEROL, L(+)-TARTARIC ACID | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-08 | | Release date: | 2013-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of an enoyl-CoA hydratase/isomerase from Xanthobacter autotrophicus Py2
TO BE PUBLISHED
|
|
