1PJ1
 
 | | RIBONUCLEOTIDE REDUCTASE R2-D84E/W48F SOAKED WITH FERROUS IONS AT PH 5 | | Descriptor: | FE (III) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Voegtli, W.C, Sommerhalter, M, Saleh, L, Baldwin, J, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-05-30 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Variable coordination geometries at the diiron(II) active site of ribonucleotide reductase R2.
J.Am.Chem.Soc., 125, 2003
|
|
7VT0
 
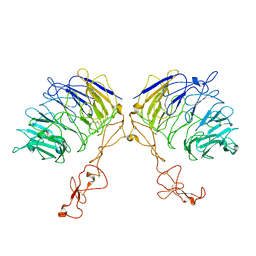 | | Dimer structure of SORLA | | Descriptor: | Sortilin-related receptor | | Authors: | Xi, Z, Cang, W, Chuang, L. | | Deposit date: | 2021-10-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal distinct apo conformations of sortilin-related receptor SORLA.
Biochem.Biophys.Res.Commun., 600, 2022
|
|
8CF5
 
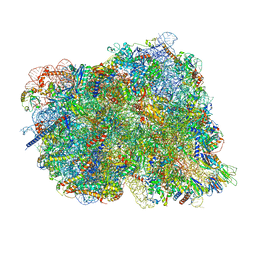 | | Translocation intermediate 1 (TI-1) of 80S S. cerevisiae ribosome with ligands and eEF2 in the presence of sordarin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-02-02 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
8CDL
 
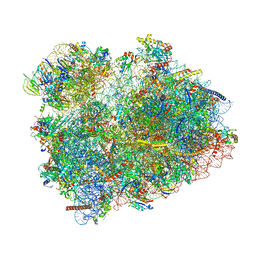 | | 80S S. cerevisiae ribosome with ligands in hybrid-2 pre-translocation (PRE-H2) complex | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-01-31 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
1PIZ
 
 | | RIBONUCLEOTIDE REDUCTASE R2 D84E MUTANT SOAKED WITH FERROUS IONS AT NEUTRAL PH | | Descriptor: | FE (III) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Voegtli, W.C, Sommerhalter, M, Saleh, L, Baldwin, J, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-05-30 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Variable coordination geometries at the diiron(II) active site of ribonucleotide reductase R2.
J.Am.Chem.Soc., 125, 2003
|
|
8CGN
 
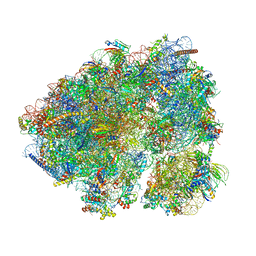 | | Non-rotated 80S S. cerevisiae ribosome with ligands | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-02-06 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
1PJG
 
 | | RNA/DNA Hybrid Decamer of CAAAGAAAAG/CTTTTCTTTG | | Descriptor: | 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3', 5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3', CALCIUM ION | | Authors: | Kopka, M.L, Lavelle, L, Han, G.W, Ng, H.-L, Dickerson, R.E. | | Deposit date: | 2003-06-02 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | An unusual sugar conformation in the structure of an RNA/DNA decamer of the polypurine tract may affect recognition by RNase H.
J.Mol.Biol., 334, 2003
|
|
8U3M
 
 | | The FARFAR-MD-NMR ensemble of an HIV-1 TAR excited state | | Descriptor: | The excited state of HIV-1 transactivation response element (31-MER) | | Authors: | Geng, A, Ganser, L, Roy, R, Shi, H, Pratihar, S, Case, D.A, Al-Hashimi, H.M. | | Deposit date: | 2023-09-07 | | Release date: | 2023-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An RNA excited conformational state at atomic resolution.
Nat Commun, 14, 2023
|
|
5LZK
 
 | | Structure of the domain of unknown function DUF1669 from human FAM83B | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, Protein FAM83B | | Authors: | Pinkas, D.M, Bufton, J.C, Williams, E.P, Shrestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-29 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.575 Å) | | Cite: | Structure of the domain of unknown function DUF1669 from human FAM83B
To Be Published
|
|
8CKU
 
 | | Translocation intermediate 1 (TI-1*) of 80S S. cerevisiae ribosome with ligands and eEF2 in the absence of sordarin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-02-16 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
1PJI
 
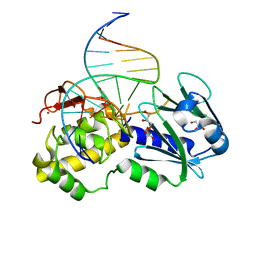 | | Crystal structure of wild type Lactococcus lactis FPG complexed to a 1,3 propanediol containing DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*TP*TP*(PDI)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Pereira, K, Serre, L, Zelwer, C, Castaing, B. | | Deposit date: | 2003-06-03 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into abasic site for Fpg specific binding and catalysis: comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA.
Nucleic Acids Res., 33, 2005
|
|
6H6S
 
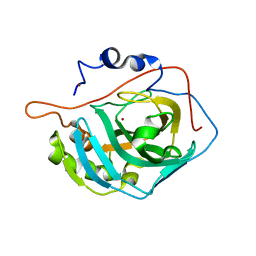 | | Sad phasing on nickel-substituted human carbonic anhydrase II | | Descriptor: | Carbonic anhydrase 2, NICKEL (II) ION | | Authors: | Calderone, V, Fragai, M, Silva, J.P, Luchinat, C, Ravera, E, Geraldes, C.F.G.C, Macedo, A.L, Cerofolini, L, Giuntini, S. | | Deposit date: | 2018-07-30 | | Release date: | 2019-01-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Non-crystallographic symmetry in proteins: Jahn-Teller-like and Butterfly-like effects?
J. Biol. Inorg. Chem., 24, 2019
|
|
8CIV
 
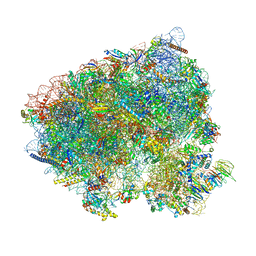 | | Translocation intermediate 5 (TI-5) of 80S S. cerevisiae ribosome with ligands and eEF2 in the presence of sordarin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
8RDS
 
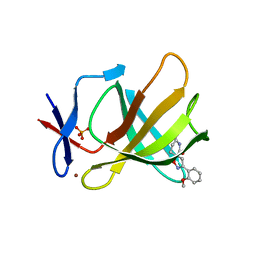 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8i | | Descriptor: | (5~{S})-3-(2-methoxyphenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
8V7W
 
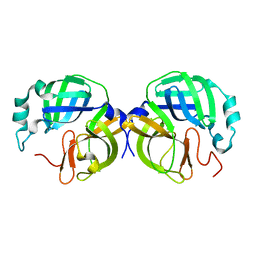 | |
5O8B
 
 | | Difference-refined excited-state structure of rsEGFP2 1ps following 400nm-laser irradiation of the off-state. | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
8RDT
 
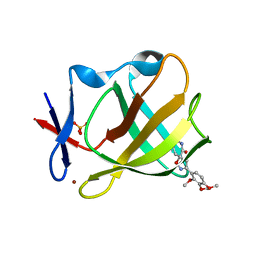 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8j | | Descriptor: | (5~{S})-3-(2,3,4-trimethoxyphenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
1FI7
 
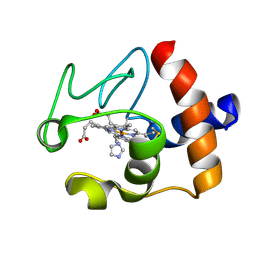 | | Solution structure of the imidazole complex of cytochrome C | | Descriptor: | CYTOCHROME C, HEME C, IMIDAZOLE | | Authors: | Banci, L, Bertini, I, Liu, G, Lu, J, Reddig, T, Tang, W, Wu, Y, Zhu, D. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Effects of extrinsic imidazole ligation on the molecular and electronic structure of cytochrome c
J.Biol.Inorg.Chem., 6, 2001
|
|
1PJ4
 
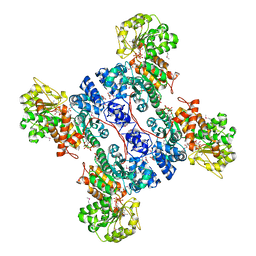 | | Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme in a pentary complex with natural substrate malate, ATP, Mn++, and allosteric activator fumarate. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MALATE, FUMARIC ACID, ... | | Authors: | Tao, X, Yang, Z, Tong, L. | | Deposit date: | 2003-05-31 | | Release date: | 2003-09-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism.
Structure, 11, 2003
|
|
5OC9
 
 | | Crystal Structure of human TMEM16K / Anoctamin 10 | | Descriptor: | (2R)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, Anoctamin-10, CALCIUM ION | | Authors: | Bushell, S.R, Pike, A.C.W, Chu, A, Tessitore, A, Rotty, B, Mukhopadhyay, S, Kupinska, K, Shrestha, L, Borkowska, O, Chalk, R, Burgess-Brown, N.A, Love, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-29 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
3URY
 
 | | Crystal Structure of Superantigen-like Protein, Exotoxin from Staphylococcus aureus subsp. aureus NCTC 8325 | | Descriptor: | CHLORIDE ION, Exotoxin | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Halavaty, A, Winsor, J, Dubrovska, I, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-22 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Superantigen-like Protein, Exotoxin from Staphylococcus aureus subsp. aureus NCTC 8325
To be Published
|
|
8RDP
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8a | | Descriptor: | (5~{S})-3-(2-chlorophenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
5O8F
 
 | | Structure of a chimaeric beta3-alpha5 GABAA receptor in complex with nanobody Nb25 and pregnanolone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit beta-3,Gamma-aminobutyric acid receptor subunit alpha-5,Gamma-aminobutyric acid receptor subunit alpha-5, Nanobody Nb25, ... | | Authors: | Miller, P.S, Scott, S, Masiulis, S, De Colibus, L, Pardon, E, Steyaert, J, Aricescu, A.R. | | Deposit date: | 2017-06-13 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for GABAA receptor potentiation by neurosteroids.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5ZIT
 
 | | Crystal structure of human Enterovirus D68 RdRp in complex with NADPH | | Descriptor: | DI(HYDROXYETHYL)ETHER, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, RdRp | | Authors: | Wang, M.L, Li, L, Chen, Y.P, Jiang, H, Zhang, Y, Su, D. | | Deposit date: | 2018-03-17 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.196 Å) | | Cite: | Structure of the enterovirus D68 RNA-dependent RNA polymerase in complex with NADPH implicates an inhibitor binding site in the RNA template tunnel.
J.Struct.Biol., 2020
|
|
1PJF
 
 | | Solid State NMR structure of the Pf1 Major Coat Protein in Magnetically Aligned Bacteriophage | | Descriptor: | COAT PROTEIN B | | Authors: | Thiriot, D.S, Nevzorov, A.A, Zagyanskiy, L, Wu, C.H, Opella, S.J. | | Deposit date: | 2003-06-02 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Structure of the coat protein in Pf1 bacteriophage determined by solid-state NMR spectroscopy.
J.Mol.Biol., 341, 2004
|
|
