4FPT
 
 | | Carbonic Anhydrase II in complex with ethyl (2Z,4R)-2-(sulfamoylimino)-1,3-thiazolidine-4-carboxylate | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Di Pizio, A, Heine, A, Klebe, G. | | Deposit date: | 2012-06-22 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | High resolution crystal structures of Carbonic Anhydrase II in complex with novel sulfamide binders
To be Published
|
|
2KVK
 
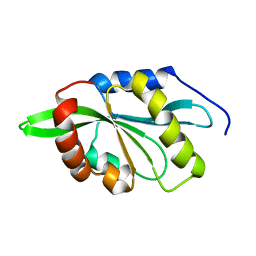 | | Solution structure of ADF/cofilin (LDCOF) from Leishmania donovani | | Descriptor: | Actin severing and dynamics regulatory protein | | Authors: | Pathak, P.P, Pulavarti, S.V, Jain, A, Sahasrabuddhe, A.A, Gupta, C.M, Arora, A. | | Deposit date: | 2010-03-16 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of ADF/cofilin from Leishmania donovani
J.Struct.Biol., 172, 2010
|
|
4BK1
 
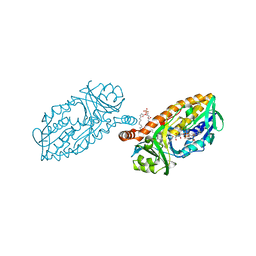 | | Crystal structure of 3-hydroxybenzoate 6-hydroxylase uncovers lipid- assisted flavoprotein strategy for regioselective aromatic hydroxylation: H213S mutant in complex with 3-hydroxybenzoate | | Descriptor: | 3-HYDROXYBENZOIC ACID, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Orru, R, Montersino, S, Barendregt, A, Westphal, A.H, van Duijn, E, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2013-04-21 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of 3-Hydroxybenzoate 6-Hydroxylase Uncovers Lipid-Assisted Flavoprotein Strategy for Regioselective Aromatic Hydroxylation
J.Biol.Chem., 288, 2013
|
|
2PMN
 
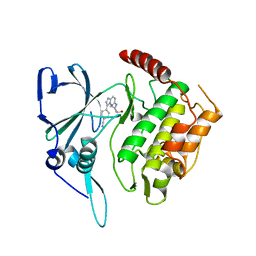 | | Crystal structure of PfPK7 in complex with an ATP-site inhibitor | | Descriptor: | 4-(6-{[(1S)-1-(HYDROXYMETHYL)-2-METHYLPROPYL]AMINO}IMIDAZO[1,2-B]PYRIDAZIN-3-YL)BENZONITRILE, Ser/Thr protein kinase, putative | | Authors: | Merckx, A, Echalier, A, Noble, M, Endicott, J. | | Deposit date: | 2007-04-23 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of P. falciparum protein kinase 7 identify an activation motif and leads for inhibitor design.
Structure, 16, 2008
|
|
4UET
 
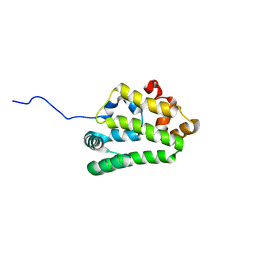 | | Diversity in the structures and ligand binding sites among the fatty acid and retinol binding proteins of nematodes revealed by Na-FAR-1 from Necator americanus | | Descriptor: | NEMATODE FATTY ACID RETINOID BINDING PROTEIN | | Authors: | Rey-Burusco, M.F, Ibanez Shimabukuro, M, Griffiths, K, Cooper, A, Kennedy, M.W, Corsico, B, Smith, B.O, Griffiths, K. | | Deposit date: | 2014-12-18 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Diversity in the Structures and Ligand Binding Sites of Nematode Fatty Acid and Retinol Binding Proteins Revealed by Na-Far-1 from Necator Americanus.
Biochem.J., 471, 2015
|
|
6UIZ
 
 | | Artificial Iron Proteins: Modelling the Active Sites in Non-Heme Dioxygenases | | Descriptor: | ACETATE ION, Streptavidin, {N-(2-{bis[(pyridin-2-yl-kappaN)methyl]amino-kappaN}ethyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}(triaza-1,2-dien-2-ium-1-ide-kappaN~1~)iron(4+) | | Authors: | Miller, K.R, Paretsky, J.D, Follmer, A.H, Heinisch, T, Mittra, K, Gul, S, Kim, I.-S, Fuller, F.D, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bhowmick, A, Sauter, N.K, Kern, J, Yano, J, Green, M.T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2019-10-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Artificial Iron Proteins: Modeling the Active Sites in Non-Heme Dioxygenases.
Inorg.Chem., 59, 2020
|
|
6UL3
 
 | |
1TJN
 
 | | Crystal structure of hypothetical protein af0721 from Archaeoglobus fulgidus | | Descriptor: | Sirohydrochlorin cobaltochelatase | | Authors: | Yin, J, Xu, X.L, Cuff, M, Walker, J.R, Edwards, A, Savchenko, A, James, M.N.G, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-06 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of af0721: a hypothetical protein bearing sequence similarity with class II chelatases in cobalamin synthesis
To be Published
|
|
4AT4
 
 | | CRYSTAL STRUCTURE OF TRKB KINASE DOMAIN IN COMPLEX WITH EX429 | | Descriptor: | 1-[4-(4-aminothieno[2,3-d]pyrimidin-5-yl)phenyl]-3-[2-fluoro-5-(trifluoromethyl)phenyl]urea, BDNF/NT-3 GROWTH FACTORS RECEPTOR | | Authors: | Bertrand, T, Kothe, M, Liu, J, Dupuy, A, Rak, A, Berne, P.F, Davis, S, Gladysheva, T, Valtre, C, Crenne, J.Y, Mathieu, M. | | Deposit date: | 2012-05-03 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | The Crystal Structures of Trka and Trkb Suggest Key Regions for Achieving Selective Inhibition.
J.Mol.Biol., 423, 2012
|
|
4AW0
 
 | | Human PDK1 Kinase Domain in Complex with Allosteric Compound PS182 Bound to the PIF-Pocket | | Descriptor: | 3-PHOSPHOINOSITIDE-DEPENDENT PROTEIN KINASE 1, ADENOSINE-5'-TRIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Schulze, J.O, Busschots, K, Lopez-Garcia, L.A, Lammi, C, Stroba, A, Zeuzem, S, Piiper, A, Alzari, P.M, Neimanis, S, Arencibia, J.M, Engel, M, Biondi, R.M. | | Deposit date: | 2012-05-30 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Substrate-Selective Inhibition of Protein Kinase Pdk1 by Small Compounds that Bind to the Pif-Pocket Allosteric Docking Site.
Chem.Biol., 19, 2012
|
|
4DYM
 
 | | Crystal structure of the ACVR1 kinase domain in complex with the imidazo[1,2-b]pyridazine inhibitor K00135 | | Descriptor: | 1-(3-{6-[(CYCLOPROPYLMETHYL)AMINO]IMIDAZO[1,2-B]PYRIDAZIN-3-YL}PHENYL)ETHANONE, Activin receptor type-1, GLYCEROL, ... | | Authors: | Chaikuad, A, Sanvitale, C, Cooper, C, Canning, P, Mahajan, P, Daga, N, Petrie, K, Alfano, I, Gileadi, O, Fedorov, O, Krojer, T, Filippakopoulos, P, Muniz, J.R.C, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-02-29 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of the ACVR1 kinase domain in complex with the imidazo[1,2-b]pyridazine inhibitor K00135
To be Published
|
|
6UI0
 
 | | Artificial Iron Proteins: Modelling the Active Sites in Non-Heme Dioxygenases | | Descriptor: | ACETATE ION, Streptavidin, {N-(2-{bis[(pyridin-2-yl-kappaN)methyl]amino-kappaN}ethyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}iron(3+) | | Authors: | Miller, K.R, Paretsky, J.D, Follmer, A.H, Heinisch, T, Mittra, K, Gul, S, Kim, I.-S, Fuller, F.D, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bhowmick, A, Sauter, N.K, Kern, J, Yano, J, Green, M.T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2019-09-29 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Artificial Iron Proteins: Modeling the Active Sites in Non-Heme Dioxygenases.
Inorg.Chem., 59, 2020
|
|
1QXX
 
 | | CRYSTAL STRUCTURE OF THE C-TERMINAL DOMAIN OF TONB | | Descriptor: | TonB protein | | Authors: | Koedding, J, Howard, P, Kaufmann, L, Polzer, P, Lustig, A, Welte, W. | | Deposit date: | 2003-09-09 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dimerization of TonB is not essential for its binding to the outer membrane siderophore receptor FhuA of Escherichia coli.
J.Biol.Chem., 279, 2004
|
|
4B8Z
 
 | | Crystal structure of human GDP-L-fucose synthase with bound NADP and GDP, rhombohedral crystal form | | Descriptor: | GDP-L-FUCOSE SYNTHASE, GUANOSINE-5'-DIPHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Vollmar, M, Krojer, T, Shafqat, N, Rojkova, A, Bunkoczi, G, Yue, W.W, Kavanagh, K, von Delft, F, Weigelt, J, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2012-08-31 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of Human Gdp-L-Fucose Synthase with Bound Nadp and Gdp, Rhombohedral Crystal Form
To be Published
|
|
4BK2
 
 | | Crystal structure of 3-hydroxybenzoate 6-hydroxylase uncovers lipid- assisted flavoprotein strategy for regioselective aromatic hydroxylation: Q301E mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATIDYLGLYCEROL-PHOSPHOGLYCEROL, PROBABLE SALICYLATE MONOOXYGENASE | | Authors: | Orru, R, Montersino, S, Barendregt, A, Westphal, A.H, van Duijn, E, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2013-04-21 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal Structure of 3-Hydroxybenzoate 6-Hydroxylase Uncovers Lipid-Assisted Flavoprotein Strategy for Regioselective Aromatic Hydroxylation
J.Biol.Chem., 288, 2013
|
|
4AT5
 
 | | CRYSTAL STRUCTURE OF TRKB KINASE DOMAIN IN COMPLEX WITH GW2580 | | Descriptor: | 5-{3-methoxy-4-[(4-methoxybenzyl)oxy]benzyl}pyrimidine-2,4-diamine, BDNF/NT-3 GROWTH FACTORS RECEPTOR, GLYCEROL | | Authors: | Bertrand, T, Kothe, M, Liu, J, Dupuy, A, Rak, A, Berne, P.F, Davis, S, Gladysheva, T, Valtre, C, Crenne, J.Y, Mathieu, M. | | Deposit date: | 2012-05-03 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The Crystal Structures of Trka and Trkb Suggest Key Regions for Achieving Selective Inhibition.
J.Mol.Biol., 423, 2012
|
|
4AVE
 
 | | C.lacteus nerve Hb in the deoxy form | | Descriptor: | ACETATE ION, GLYCEROL, NEURAL HEMOGLOBIN, ... | | Authors: | Germani, F, Pesce, A, Venturini, A, Moens, L, Bolognesi, M, Dewilde, S, Nardini, M. | | Deposit date: | 2012-05-25 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High Resolution Crystal Structures of the Cerebratulus Lacteus Mini-Hb in the Unligated and Carbomonoxy States.
Int.J.Mol.Sci., 13, 2012
|
|
4AW1
 
 | | Human PDK1 Kinase Domain in Complex with Allosteric Compound PS210 Bound to the PIF-Pocket | | Descriptor: | 3-PHOSPHOINOSITIDE-DEPENDENT PROTEIN KINASE 1, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Schulze, J.O, Busschots, K, Lopez-Garcia, L.A, Lammi, C, Stroba, A, Zeuzem, S, Piiper, A, Alzari, P.M, Neimanis, S, Arencibia, J.M, Engel, M, Biondi, R.M. | | Deposit date: | 2012-05-30 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Substrate-Selective Inhibition of Protein Kinase Pdk1 by Small Compounds that Bind to the Pif-Pocket Allosteric Docking Site.
Chem.Biol., 19, 2012
|
|
4EEY
 
 | | Crystal structure of human DNA polymerase eta in ternary complex with a cisplatin DNA adduct | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*TP*TP*GP*GP*TP*CP*TP*CP*CP*TP*CP*C)-3', 5'-D(*TP*GP*GP*AP*GP*GP*AP*GP*A)-3', ... | | Authors: | Ummat, A, Rechkoblit, O, Jain, R, Choudhury, J.R, Johnson, R.E, Silverstein, T.D, Buku, A, Lone, S, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural basis for cisplatin DNA damage tolerance by human polymerase {eta} during cancer chemotherapy.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4ASZ
 
 | | Crystal structure of apo TrkB kinase domain | | Descriptor: | BDNF/NT-3 GROWTH FACTORS RECEPTOR | | Authors: | Bertrand, T, Kothe, M, Liu, J, Dupuy, A, Rak, A, Berne, P.F, Davis, S, Gladysheva, T, Valtre, C, Crenne, J.Y, Mathieu, M. | | Deposit date: | 2012-05-03 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structures of Trka and Trkb Suggest Key Regions for Achieving Selective Inhibition.
J.Mol.Biol., 423, 2012
|
|
1QBJ
 
 | | CRYSTAL STRUCTURE OF THE ZALPHA Z-DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*G)-3'), PROTEIN (DOUBLE-STRANDED RNA SPECIFIC ADENOSINE DEAMINASE (ADAR1)) | | Authors: | Schwartz, T, Rould, M.A, Rich, A. | | Deposit date: | 1999-04-22 | | Release date: | 1999-07-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA.
Science, 284, 1999
|
|
4BA6
 
 | | High Resolution structure of the C-terminal family 65 Carbohydrate Binding Module (CBM65B) of endoglucanase Cel5A from Eubacterium cellulosolvens | | Descriptor: | Endoglucanase cel5A, GLYCEROL | | Authors: | Venditto, I, Luis, A.S, Basle, A, Temple, M, Ferreira, L.M.A, Fontes, C.M.G.A, Gilbert, H.J, Najmudin, S. | | Deposit date: | 2012-09-11 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Understanding how noncatalytic carbohydrate binding modules can display specificity for xyloglucan.
J. Biol. Chem., 288, 2013
|
|
3SR3
 
 | | Crystal structure of the w180a mutant of microcin immunity protein mccf from Bacillus anthracis shows the active site loop in the open conformation. | | Descriptor: | Microcin immunity protein MccF | | Authors: | Nocek, B, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-06 | | Release date: | 2011-08-10 | | Last modified: | 2013-01-09 | | Method: | X-RAY DIFFRACTION (1.495 Å) | | Cite: | Structural and functional characterization of microcin C resistance peptidase MccF from Bacillus anthracis.
J.Mol.Biol., 420, 2012
|
|
4EOJ
 
 | | Thr 160 phosphorylated CDK2 H84S, Q85M, K89D - human cyclin A3 complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Echalier, A, Cot, E, Camasses, A, Hodimont, E, Hoh, F, Sheinerman, F, Krasinska, L, Fisher, D. | | Deposit date: | 2012-04-14 | | Release date: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An integrated chemical biology approach provides insight into Cdk2 functional redundancy and inhibitor sensitivity.
Chem.Biol., 19, 2012
|
|
1URI
 
 | | AZURIN MUTANT WITH MET 121 REPLACED BY GLN | | Descriptor: | AZURIN, COPPER (II) ION, SULFATE ION | | Authors: | Romero, A, Nar, H, Huber, R, Messerschmidt, A. | | Deposit date: | 1996-11-14 | | Release date: | 1997-04-01 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | X-ray analysis and spectroscopic characterization of M121Q azurin. A copper site model for stellacyanin.
J.Mol.Biol., 229, 1993
|
|
