2V1Y
 
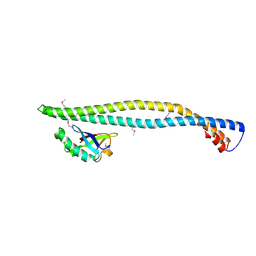 | | Structure of a phosphoinositide 3-kinase alpha adaptor-binding domain (ABD) in a complex with the iSH2 domain from p85 alpha | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY SUBUNIT ALPHA, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT ALPHA ISOFORM | | Authors: | Miled, N, Yan, Y, Hon, W.C, Perisic, O, Zvelebil, M, Inbar, Y, Schneidman-Duhovny, D, Wolfson, H.J, Backer, J.M, Williams, R.L. | | Deposit date: | 2007-05-30 | | Release date: | 2007-07-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of Two Classes of Cancer Mutations in the Phosphoinositide 3-Kinase Catalytic Subunit.
Science, 317, 2007
|
|
1R6Q
 
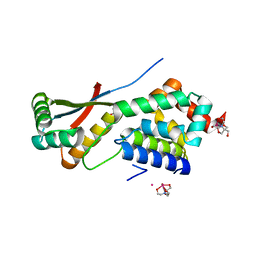 | | ClpNS with fragments | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpA, ATP-dependent Clp protease adaptor protein clpS, BIS-(2-HYDROXYETHYL)AMINO-TRIS(HYDROXYMETHYL)METHANE YTTRIUM, ... | | Authors: | Xia, D, Maurizi, M.R, Guo, F, Singh, S.K, Esser, L. | | Deposit date: | 2003-10-16 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystallographic investigation of peptide binding sites in the N-domain of the ClpA chaperone.
J.Struct.Biol., 146, 2004
|
|
1U02
 
 | | Crystal structure of trehalose-6-phosphate phosphatase related protein | | Descriptor: | GLYCEROL, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Krishnamurthy, N.R, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-12 | | Release date: | 2004-07-20 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of trehalose-6-phosphate phosphatase-related protein: biochemical and biological implications.
Protein Sci., 15, 2006
|
|
2V3E
 
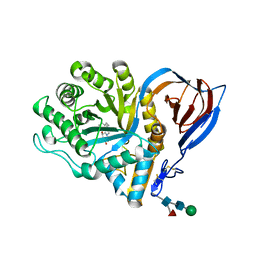 | | acid-beta-glucosidase with N-nonyl-deoxynojirimycin | | Descriptor: | (2R,3R,4R,5S)-2-(HYDROXYMETHYL)-1-NONYLPIPERIDINE-3,4,5-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUCOSYLCERAMIDASE, ... | | Authors: | Brumshtein, B, Greenblatt, H.M, Butters, T.D, Shaaltiel, Y, Aviezer, D, Silman, I, Futerman, A.H, Sussman, J.L. | | Deposit date: | 2007-06-17 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Complexes of N-Butyl- and N-Nonyl-Deoxynojirimycin Bound to Acid Beta-Glucosidase: Insights Into the Mechanism of Chemical Chaperone Action in Gaucher Disease.
J.Biol.Chem., 282, 2007
|
|
2VIN
 
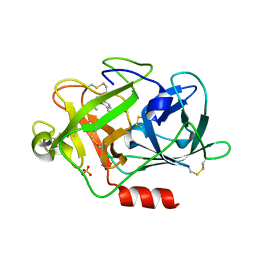 | | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator | | Descriptor: | (2R)-1-(2,6-dimethylphenoxy)propan-2-amine, ACETATE ION, SULFATE ION, ... | | Authors: | Frederickson, M, Callaghan, O, Chessari, G, Congreve, M, Cowan, S.R, Matthews, J.E, McMenamin, R, Smith, D, Vinkovic, M, Wallis, N.G. | | Deposit date: | 2007-12-05 | | Release date: | 2008-01-22 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator
J.Med.Chem., 51, 2008
|
|
1U0D
 
 | | Y33H Mutant of Homing endonuclease I-CreI | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*TP*CP*GP*C)-3', 5'-D(*GP*CP*GP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*CP*CP*G)-3', DNA endonuclease I-CreI | | Authors: | Sussman, D, Chadsey, M, Fauce, S, Engel, A, Bruett, A, Monnat, R, Stoddard, B.L, Seligman, L.M. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Isolation and characterization of new homing endonuclease specificities at individual target site positions.
J.Mol.Biol., 342, 2004
|
|
2VA9
 
 | | Structure of native TcAChE after a 9 seconds annealing to room temperature during the first 5 seconds of which laser irradiation at 266nm took place | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE | | Authors: | Colletier, J.-P, Sanson, B, Royant, A, Specht, A, Nachon, F, Masson, P, Zaccai, G, Sussman, J.L, Goeldner, M, Silman, I, Bourgeois, D, Weik, M. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Use of a 'Caged' Analog to Study Traffic of Choline within Acetylcholinesterase by Kinetic Crystallography
Acta Crystallogr.,Sect.D, 63, 2007
|
|
7OLZ
 
 | | Crystal structure of the SARS-CoV-2 RBD with neutralizing-VHHs Re5D06 and Re9F06 | | Descriptor: | 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, Nanobody Re5D06, Nanobody Re9F06, ... | | Authors: | Aksu, M, Guttler, T, Gorlich, D. | | Deposit date: | 2021-05-20 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Neutralization of SARS-CoV-2 by highly potent, hyperthermostable, and mutation-tolerant nanobodies.
Embo J., 40, 2021
|
|
7P54
 
 | | Cryo-EM structure of human TTYH2 in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 2 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
7P5J
 
 | | Cryo-EM structure of human TTYH1 in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 1 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
7OS8
 
 | | NMR SOLUTION STRUCTURE OF [Pro3,DLeu9]TL | | Descriptor: | PHE-VAL-PRO-TRP-PHE-SER-LYS-PHE-DLE-GLY-ARG-ILE-LEU-NH2 | | Authors: | Brancaccio, D, Carotenuto, A. | | Deposit date: | 2021-06-08 | | Release date: | 2021-08-11 | | Last modified: | 2021-08-25 | | Method: | SOLUTION NMR | | Cite: | First-in-Class Cyclic Temporin L Analogue: Design, Synthesis, and Antimicrobial Assessment.
J.Med.Chem., 64, 2021
|
|
7ON5
 
 | | Crystal structure of the SARS-CoV-2 neutralizing nanobody Re5D06 | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Nanobody Re5D06 | | Authors: | Aksu, M, Guttler, T, Gorlich, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Neutralization of SARS-CoV-2 by highly potent, hyperthermostable, and mutation-tolerant nanobodies.
Embo J., 40, 2021
|
|
2VFY
 
 | |
2VH0
 
 | | Structure and property based design of factor Xa inhibitors:biaryl pyrrolidin-2-ones incorporating basic heterocyclic motifs | | Descriptor: | 2-(5-chlorothiophen-2-yl)-N-[(3S)-1-(4-{2-[(dimethylamino)methyl]-1H-imidazol-1-yl}-2-fluorophenyl)-2-oxopyrrolidin-3-yl]ethanesulfonamide, ACTIVATED FACTOR XA HEAVY CHAIN, ACTIVATED FACTOR XA LIGHT CHAIN, ... | | Authors: | Young, R.J, Borthwick, A.D, Brown, D, Burns-Kurtis, C.L, Campbell, M, Chan, C, Charbaut, M, Convery, M.A, Diallo, H, Hortense, E, Irving, W.R, Kelly, H.A, King, N.P, Kleanthous, S, Mason, A.M, Pateman, A.J, Patikis, A, Pinto, I.L, Pollard, D.R, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E, Zhou, P. | | Deposit date: | 2007-11-16 | | Release date: | 2008-11-25 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and property based design of factor Xa inhibitors: biaryl pyrrolidin-2-ones incorporating basic heterocyclic motifs.
Bioorg. Med. Chem. Lett., 18, 2008
|
|
5M3A
 
 | | Crystal structure of BRD4 BROMODOMAIN 1 IN COMPLEX WITH LIGAND 2 | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-6-(1-methyl-5-phenoxy-pyrazol-4-yl)-[1,2,4]triazolo[4,3-b]pyridazine, Bromodomain-containing protein 4 | | Authors: | Kessler, D, Mayer, M, Engelhardt, H, Wolkerstorfer, B, Geist, L. | | Deposit date: | 2016-10-14 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Direct NMR Probing of Hydration Shells of Protein Ligand Interfaces and Its Application to Drug Design.
J. Med. Chem., 60, 2017
|
|
2VLD
 
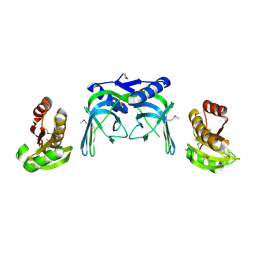 | | crystal structure of a repair endonuclease from Pyrococcus abyssi | | Descriptor: | Endonuclease NucS | | Authors: | Ren, B, Kuhn, J, Meslet-Cladiere, L, Briffotaux, J, Norais, C, Lavigne, R, Flament, D, Ladenstein, R, Myllykallio, H. | | Deposit date: | 2008-01-14 | | Release date: | 2009-05-19 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and function of a novel endonuclease acting on branched DNA substrates.
EMBO J., 28, 2009
|
|
5LRU
 
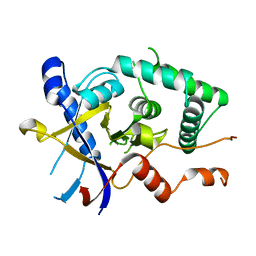 | | Structure of Cezanne/OTUD7B OTU domain | | Descriptor: | OTU domain-containing protein 7B | | Authors: | Mevissen, T.E.T, Kulathu, Y, Mulder, M.P.C, Geurink, P.P, Maslen, S.L, Gersch, M, Elliott, P.R, Burke, J.E, van Tol, B.D.M, Akutsu, M, El Oualid, F, Kawasaki, M, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2016-08-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne.
Nature, 538, 2016
|
|
1N8M
 
 | | Solution structure of Pi4, a four disulfide bridged scorpion toxin active on potassium channels | | Descriptor: | Potassium channel blocking toxin 4 | | Authors: | Guijarro, J.I, M'Barek, S, Olamendi-Portugal, T, Gomez-Lagunas, F, Garnier, D, Rochat, H, Possani, L.D, Sabatier, J.M, Delepierre, M. | | Deposit date: | 2002-11-21 | | Release date: | 2003-09-02 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Pi4, a short four-disulfide-bridged scorpion toxin specific of potassium channels.
Protein Sci., 12, 2003
|
|
1U5H
 
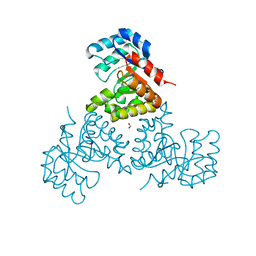 | | Structure of Citrate Lyase beta subunit from Mycobacterium tuberculosis | | Descriptor: | FORMIC ACID, citE | | Authors: | Goulding, C.W, Lekin, T, Kim, C.Y, Segelke, B, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-07-27 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure and computational analysis of Mycobacterium tuberculosis protein CitE suggest a novel enzymatic function.
J.Mol.Biol., 365, 2007
|
|
5M37
 
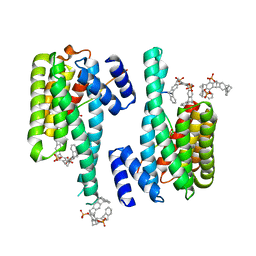 | | The molecular tweezer CLR01 stabilizes a disordered protein-protein interface | | Descriptor: | (1R,5S,9S,16R,20R,24S,28S,35R)-3,22-Bis(dihydroxyphosphoryloxy)tridecacyclo[22.14.1.15,20.19,16.128,35.02,23.04,21.06,19.08,17.010,15.025,38.027,36.029,34]dotetraconta-2(23),3,6,8(17),10,12,14,18,21,25,27(36),29,31,33,37-pentadecaene, 14-3-3 protein zeta/delta, BENZOIC ACID, ... | | Authors: | Bier, D, Ottmann, C. | | Deposit date: | 2016-10-14 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Molecular Tweezer CLR01 Stabilizes a Disordered Protein-Protein Interface.
J. Am. Chem. Soc., 139, 2017
|
|
5NRW
 
 | |
7P4Z
 
 | |
7P1W
 
 | |
7OYG
 
 | | Dimeric form of SARS-CoV-2 RNA-dependent RNA polymerase | | Descriptor: | RNA (5'-R(P*CP*UP*AP*CP*GP*CP*AP*GP*UP*G)-3'), RNA (5'-R(P*UP*GP*CP*AP*CP*UP*GP*CP*GP*UP*AP*G)-3'), SARS-CoV-2 RNA-dependent RNA polymerase (nsp12), ... | | Authors: | Jochheim, F.A, Tegunov, D, Hillen, H.S, Schmitzova, J, Kokic, G, Dienemann, C, Cramer, P. | | Deposit date: | 2021-06-24 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | The structure of a dimeric form of SARS-CoV-2 polymerase
Communications Biology, 4, 2021
|
|
2V4B
 
 | | Crystal Structure of Human ADAMTS-1 catalytic Domain and Cysteine- Rich Domain (apo-form) | | Descriptor: | ADAMTS-1, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Gerhardt, S, Hassall, G, Hawtin, P, McCall, E, Flavell, L, Minshull, C, Hargreaves, D, Ting, A, Pauptit, R.A, Parker, A.E, Abbott, W.M. | | Deposit date: | 2007-06-28 | | Release date: | 2008-01-15 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Human Adamts-1 Reveal a Conserved Catalytic Domain and a Disintegrin-Like Domain with a Fold Homologous to Cysteine-Rich Domains.
J.Mol.Biol., 373, 2007
|
|
