6QM9
 
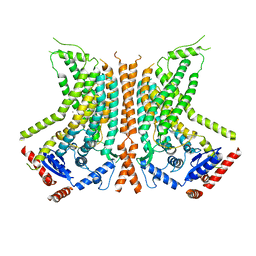 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in nanodisc (open state) | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QM4
 
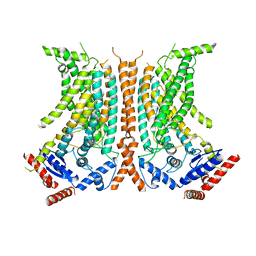 | | Cryo-EM structure of calcium-free nhTMEM16 lipid scramblase in nanodisc | | Descriptor: | Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QM6
 
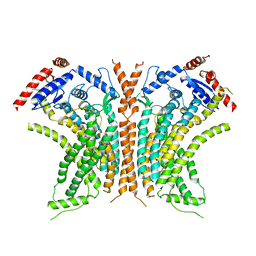 | | Cryo-EM structure of calcium-free nhTMEM16 lipid scramblase in DDM | | Descriptor: | Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
8Q9R
 
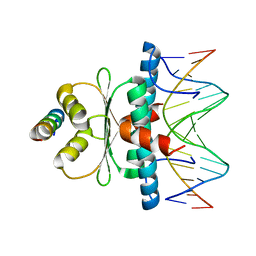 | | Crystal structure of MADS-box/MEF2D N-terminal domain bound to dsDNA and HDAC9 deacetylase binding motif | | Descriptor: | Histone deacetylase 9 (HDAC9) binding motif peptide: EVKQKLQEFLLSKS, MADS box dsDNA: AACTATTTATAAGA, MADS box dsDNA: TCTTATAAATAGT, ... | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzoccato, Y, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-08-20 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
6QMA
 
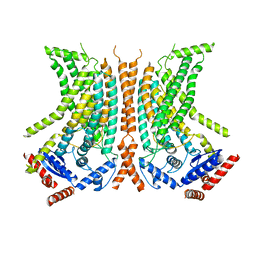 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in nanodisc (intermediate state) | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
8Q9P
 
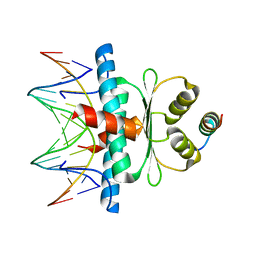 | | Crystal Structure of the MADS-box/MEF2 Domain of MEF2D bound to dsDNA and HDAC5 deacetylase binding motif | | Descriptor: | HDAC5 (histone deacetylase 5) binding motif peptide: TRP-GLY-SER-GLY-GLU-VAL-LYS-LEU-ARG-LEU-GLN-GLU-PHE-LEU-LEU-SER-LYS-SER, MADS box dsDNA fw: AACTATTTATAAGA, MADS box dsNA rev:TCTTATAAATAGTT, ... | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzocato, Y, Biondi, B, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-08-20 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
8Q9Q
 
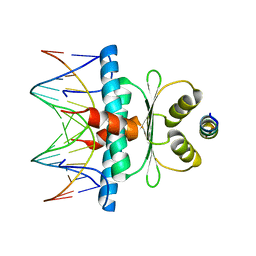 | | Crystal Structure of the MADS-box/MEF2 Domain of MEF2D bound to dsDNA and HDAC7 deacetylase binding motif | | Descriptor: | HDAC7 (histone deacetylase 7) binding motif peptide: GLY-VAL-VAL-LYS-GLN-LYS-LEU-ALA-GLU-VAL-ILE-LEU-LYS-LYS-GLN, MADS box dsDNA: AACTATTTATAAGA, MADS box dsDNA: TCTTATAAATAGTT, ... | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzocato, Y, Biondi, B, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-08-20 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
6QM5
 
 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in DDM | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
8Q9N
 
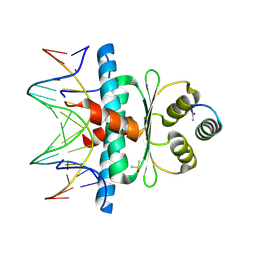 | | Crystal Structure of the MADS-box/MEF2 Domain of MEF2D bound to dsDNA and MITR deacetylase binding motif mutant L151V. | | Descriptor: | DIMETHYL SULFOXIDE, DNA MADS box, MEF2D protein, ... | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzocato, Y, Biondi, B, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-08-20 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
1A1R
 
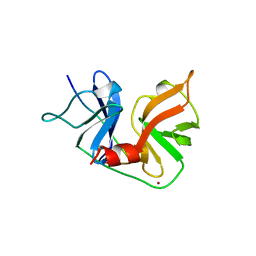 | | HCV NS3 PROTEASE DOMAIN:NS4A PEPTIDE COMPLEX | | Descriptor: | NS3 PROTEIN, NS4A PROTEIN, ZINC ION | | Authors: | Kim, J.L, Morgenstern, K.A, Lin, C, Fox, T, Dwyer, M.D, Landro, J.A, Chambers, S.P, Markland, W, Lepre, C.A, O'Malley, E.T, Harbeson, S.L, Rice, C.M, Murcko, M.A, Caron, P.R, Thomson, J.A. | | Deposit date: | 1997-12-15 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide.
Cell(Cambridge,Mass.), 87, 1996
|
|
8QRQ
 
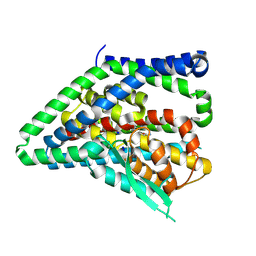 | | ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS.2) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRW
 
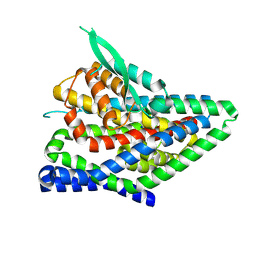 | | ASCT2 protomer in lipid nanodiscs under low Na+ concentration in the intermediate outward-facing state (iOFS-up) | | Descriptor: | Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRR
 
 | | ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS.3) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRU
 
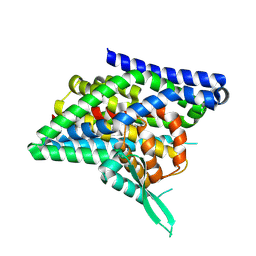 | | ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the intermediate outward-facing state (iOFS-down) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0) | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRP
 
 | | ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS.1) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRS
 
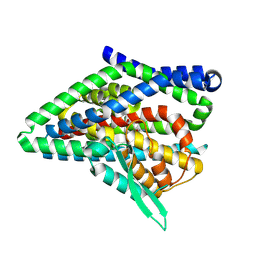 | | ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the intermediate outward-facing state (iOFS-up) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0) | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRV
 
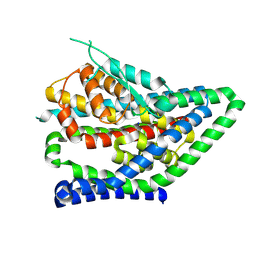 | | ASCT2 protomer in lipid nanodiscs under low Na+ concentration in the outward-facing state (OFS) | | Descriptor: | Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRO
 
 | | ASCT2 trimer in lipid nanodiscs with bound glutamine and Na+ ions in the outward-facing state (OFS) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
7VVP
 
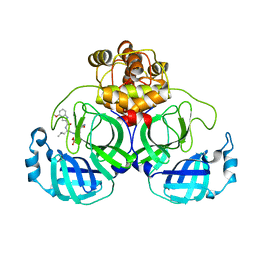 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2021-11-07 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
7WQH
 
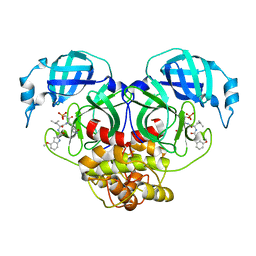 | | Crystal structure of HCoV-NL63 main protease with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zhong, F.L, Zhou, X.L, Lin, C, Zeng, P, Li, J, Zhang, J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
7XLT
 
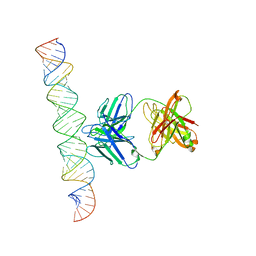 | | Cryo-EM Structure of R-loop monoclonal antibody S9.6 in recognizing RNA:DNA hybrids | | Descriptor: | DNA, RNA, S9.6 Fab HC, ... | | Authors: | Li, Q, Lin, C, Luo, Z, Li, H, Li, X, Sun, Q. | | Deposit date: | 2022-04-22 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of R-loop monoclonal antibody S9.6 in recognizing RNA:DNA hybrids.
J Genet Genomics, 49, 2022
|
|
7VLQ
 
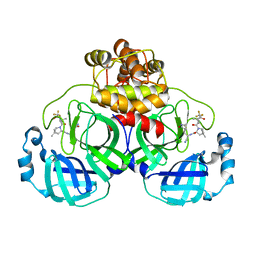 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07321332 in spacegroup P212121 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zhang, J, Li, J. | | Deposit date: | 2021-10-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.939106 Å) | | Cite: | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VLP
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07321332 in spacegroup P1211 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, Replicase polyprotein 1a | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Li, J, Zhang, J. | | Deposit date: | 2021-10-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.50251937 Å) | | Cite: | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
1A1V
 
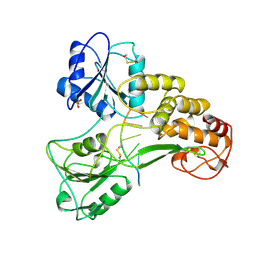 | | HEPATITIS C VIRUS NS3 HELICASE DOMAIN COMPLEXED WITH SINGLE STRANDED SDNA | | Descriptor: | DNA (5'-D(*UP*UP*UP*UP*UP*UP*UP*U)-3'), PROTEIN (NS3 PROTEIN), SULFATE ION | | Authors: | Kim, J.L, Morgenstern, K.A, Griffith, J.P, Dwyer, M.D, Thomson, J.A, Murcko, M.A, Lin, C, Caron, P.R. | | Deposit date: | 1997-12-17 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding.
Structure, 6, 1998
|
|
7XRS
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with inhibitor YH-53 | | Descriptor: | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, Replicase polyprotein 1a | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-05-11 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
