4QSX
 
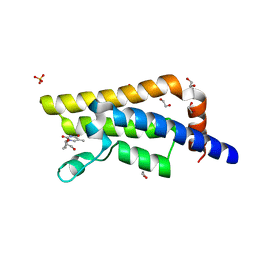 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) in complex with 3'-deoxy thymidine | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2R,5S)-5-(hydroxymethyl)tetrahydrofuran-2-yl]-5-methylpyrimidine-2,4(1H,3H)-dione, ATPase family AAA domain-containing protein 2, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
6Z54
 
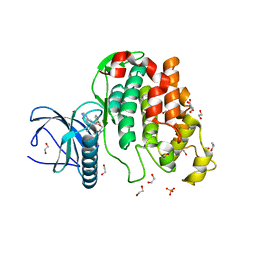 | | Crystal structure of CLK3 in complex with macrocycle ODS2003178 | | Descriptor: | 1,2-ETHANEDIOL, 11,15-Dimethyl-6-(oxan-4-yloxy)-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, Dual specificity protein kinase CLK3, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of CLK3 in complex with macrocycle ODS2003178
To Be Published
|
|
6Z57
 
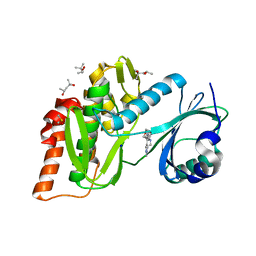 | | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2004078 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 10-(2-morpholin-4-ylethyl)-7-oxa-10,13,17,18,21-pentazatetracyclo[12.5.2.12,6.017,20]docosa-1(20),2(22),3,5,14(21),15,18-heptaene, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2004078
To Be Published
|
|
6Z53
 
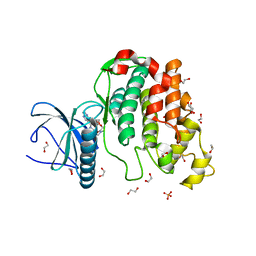 | | Crystal structure of CLK3 in complex with macrocycle ODS2003128 | | Descriptor: | 1,2-ETHANEDIOL, 6-(2-methoxyethoxy)-11,15-dimethyl-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, Dual specificity protein kinase CLK3, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of CLK3 in complex with macrocycle ODS2003128
To Be Published
|
|
6Z58
 
 | | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2003791 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8,14,18,19,22-pentazatetracyclo[13.5.2.12,6.018,21]tricosa-1(21),2,4,6(23),15(22),16,19-heptaen-7-one, SODIUM ION, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2003791
To Be Published
|
|
6Z50
 
 | | Crystal structure of CLK1 in complex with macrocycle ODS2003208 | | Descriptor: | 1,2-ETHANEDIOL, 11,15-dimethyl-6-(oxan-4-yloxy)-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, Dual specificity protein kinase CLK1, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of CLK1 in complex with macrocycle ODS2003208
To Be Published
|
|
6Z5B
 
 | | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2003128 | | Descriptor: | 6-(2-methoxyethoxy)-11,15-dimethyl-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, DIMETHYL SULFOXIDE, Serine/threonine-protein kinase haspin | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2003128
To Be Published
|
|
6Z5C
 
 | | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2004070 | | Descriptor: | 7,10-Dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.12,6.017,20]docosa-1(20),2(22),3,5,14(21),15,18-heptaene-5-carboxylic acid, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2004070
To Be Published
|
|
6Z4Z
 
 | | Crystal structure of CLK1 in complex with macrocycle ODS2004070 | | Descriptor: | 7,10-Dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.12,6.017,20]docosa-1(20),2(22),3,5,14(21),15,18-heptaene-5-carboxylic acid, Dual specificity protein kinase CLK1, GLYCEROL, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of CLK1 in complex with macrocycle ODS2004070
To Be Published
|
|
6Z5D
 
 | | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2004082 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 11-methyl-8,11,14,18,19,22-hexazatetracyclo[13.5.2.12,6.018,21]tricosa-1(21),2(23),3,5,15(22),16,19-heptaen-7-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2004082
To Be Published
|
|
6Z55
 
 | | Crystal structure of CLK3 in complex with macrocycle ODS2004070 | | Descriptor: | 1,2-ETHANEDIOL, 7,10-Dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.12,6.017,20]docosa-1(20),2(22),3,5,14(21),15,18-heptaene-5-carboxylic acid, Dual specificity protein kinase CLK3, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of CLK3 in complex with macrocycle ODS2004070
To Be Published
|
|
6Z51
 
 | | Crystal structure of CLK3 in complex with macrocycle ODS2002941 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein kinase CLK3, N-cyclopentyl-2-[(11,15-dimethyl-10-oxo-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-6-yl)oxy]acetamide, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of CLK3 in complex with macrocycle ODS2002941
To Be Published
|
|
6Z5A
 
 | | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2002941 | | Descriptor: | DIMETHYL SULFOXIDE, N-cyclopentyl-2-[(11,15-dimethyl-10-oxo-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-6-yl)oxy]acetamide, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2002941
To Be Published
|
|
7B2S
 
 | |
7B2R
 
 | |
7A77
 
 | | Crystal structure of RXR alpha LBD in complexes with palmitic acid and GRIP-1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Nuclear receptor coactivator 2, ... | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comprehensive Set of Tertiary Complex Structures and Palmitic Acid Binding Provide Molecular Insights into Ligand Design for RXR Isoforms.
Int J Mol Sci, 21, 2020
|
|
7A79
 
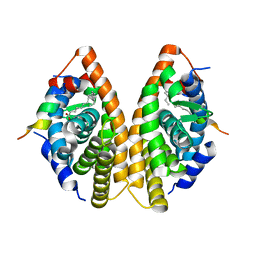 | | Crystal structure of RXR gamma LBD in complexes with palmitic acid and GRIP-1 peptide | | Descriptor: | Nuclear receptor coactivator 2, PALMITIC ACID, Retinoic acid receptor RXR-gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Comprehensive Set of Tertiary Complex Structures and Palmitic Acid Binding Provide Molecular Insights into Ligand Design for RXR Isoforms.
Int J Mol Sci, 21, 2020
|
|
7A78
 
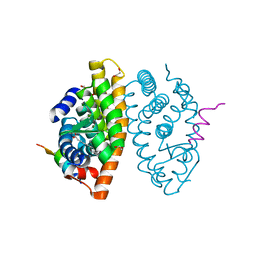 | | Crystal structure of RXR beta LBD in complexes with palmitic acid and GRIP-1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Nuclear receptor coactivator 2, ... | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Comprehensive Set of Tertiary Complex Structures and Palmitic Acid Binding Provide Molecular Insights into Ligand Design for RXR Isoforms.
Int J Mol Sci, 21, 2020
|
|
8PP0
 
 | | Crystal structure of Retinoic Acid Receptor alpha (RXRA) in complexed with JP147 | | Descriptor: | 3-[4-[2,3-dihydro-1H-inden-4-yl(methyl)amino]-6-(trifluoromethyl)pyrimidin-2-yl]oxypropanoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Chaikuad, A, Pollinger, J, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-07-05 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of a Highly Potent Partial RXR Agonist with Superior Physicochemical Properties.
J.Med.Chem., 67, 2024
|
|
9FLC
 
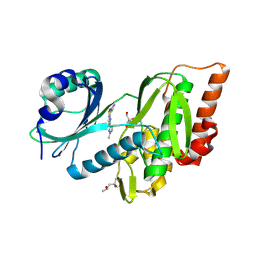 | | Crystal structure of haspin (GSG2) in complex with MU1668 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(1-methylpyrazol-3-yl)-3-pyridin-4-yl-thieno[3,2-b]pyridine, GLYCEROL, ... | | Authors: | Chaikuad, A, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-06-04 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Thieno[3,2-b]pyridine: Attractive scaffold for highly selective inhibitors of underexplored protein kinases with variable binding mode.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9FLB
 
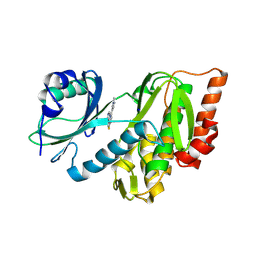 | | Crystal structure of haspin (GSG2) in complex with MU1464 | | Descriptor: | 5-(1-methylpyrazol-4-yl)-3-pyridin-4-yl-thieno[3,2-b]pyridine, SODIUM ION, Serine/threonine-protein kinase haspin | | Authors: | Chaikuad, A, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-06-04 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Thieno[3,2-b]pyridine: Attractive scaffold for highly selective inhibitors of underexplored protein kinases with variable binding mode.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9GLQ
 
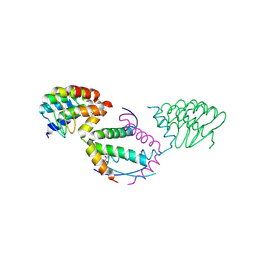 | | Crystal structure of p73 tetramerisation domain in complex with darpins 1800 | | Descriptor: | COBALT (II) ION, Darpins 1800, GLYCEROL, ... | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-08-27 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DARPins as a novel tool to detect and degrade p73.
Cell Death Dis, 15, 2024
|
|
8P9E
 
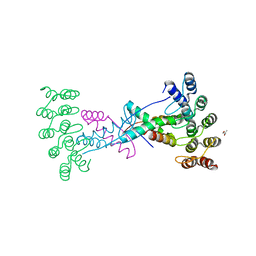 | | Crystal structure of wild type p63-p73 heterotetramer (tetramerisation domain) in complex with darpin 1810 F11 | | Descriptor: | Darpin 1810 F11, GLYCEROL, Isoform 2 of Tumor protein 63, ... | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-05 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | DARPins detect the formation of hetero-tetramers of p63 and p73 in epithelial tissues and in squamous cell carcinoma.
Cell Death Dis, 14, 2023
|
|
8P9C
 
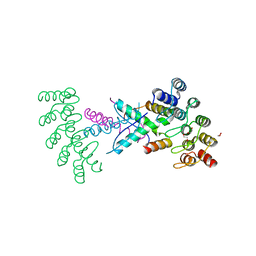 | | Crystal structure of p63-p73 heterotetramer (tetramerisation domain) in complex with darpin 1810 F11 | | Descriptor: | 1,2-ETHANEDIOL, Darpin 1810 F11, Tumor protein 63, ... | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-05 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | DARPins detect the formation of hetero-tetramers of p63 and p73 in epithelial tissues and in squamous cell carcinoma.
Cell Death Dis, 14, 2023
|
|
8P9D
 
 | | Crystal structure of p63-p73 heterotetramer (tetramerisation domain) in complex with darpin 1810 A2 | | Descriptor: | Darpin 1810 A2, Tumor protein 63, Tumor protein p73 | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-05 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | DARPins detect the formation of hetero-tetramers of p63 and p73 in epithelial tissues and in squamous cell carcinoma.
Cell Death Dis, 14, 2023
|
|
