8Q9D
 
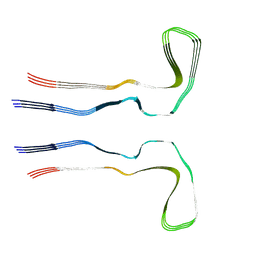 | |
8Q8R
 
 | | Tau - AD-PHF | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Lovestam, S, Li, D, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-08-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Disease-specific tau filaments assemble via polymorphic intermediates.
Nature, 625, 2024
|
|
8Q99
 
 | |
8Q9H
 
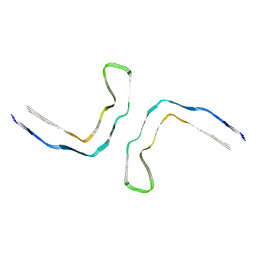 | |
8Q8F
 
 | |
8Q8S
 
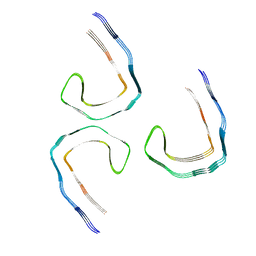 | | Tau - AD-THF | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Lovestam, S, Li, D, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-08-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Disease-specific tau filaments assemble via polymorphic intermediates.
Nature, 625, 2024
|
|
8Q8Z
 
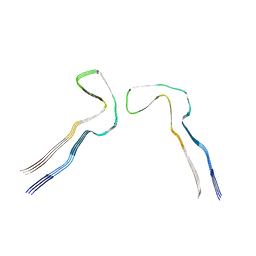 | |
8Q8L
 
 | |
8Q8U
 
 | |
8Q8W
 
 | |
8Q9A
 
 | |
8Q8E
 
 | |
8Q8Y
 
 | |
8Q9E
 
 | |
8Q8X
 
 | |
8Q9B
 
 | |
8Q8V
 
 | | Tau - CTE-MIA3 | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Lovestam, S, Li, D, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-08-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Disease-specific tau filaments assemble via polymorphic intermediates.
Nature, 625, 2024
|
|
8Q9C
 
 | |
8PPO
 
 | |
5ZGL
 
 | | hnRNP A1 segment GGGYGGS (residues 234-240) | | Descriptor: | 7-mer peptide from Heterogeneous nuclear ribonucleoprotein A1 | | Authors: | Xie, M, Luo, F, Gui, X, Zhao, M, He, J, Li, D, Liu, C. | | Deposit date: | 2018-03-09 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
5ZGD
 
 | | hnRNPA1 reversible amyloid core GFGGNDNFG (residues 209-217) determined by X-ray | | Descriptor: | GLY-PHE-GLY-GLY-ASN-ASP-ASN-PHE-GLY | | Authors: | Gui, X, Xie, M, Zhao, M, Luo, F, He, J, Li, D, Liu, C. | | Deposit date: | 2018-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
4NC3
 
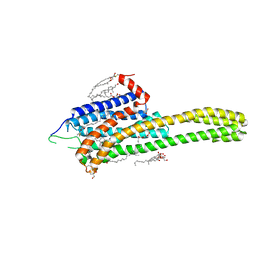 | | Crystal structure of the 5-HT2B receptor solved using serial femtosecond crystallography in lipidic cubic phase. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHOLESTEROL, ... | | Authors: | Liu, W, Wacker, D, Gati, C, Han, G.W, James, D, Wang, D, Nelson, G, Weierstall, U, Katritch, V, Barty, A, Zatsepin, N.A, Li, D, Messerschmidt, M, Boutet, S, Williams, G.J, Koglin, J.E, Seibert, M.M, Wang, C, Shah, S.T.A, Basu, S, Fromme, R, Kupitz, C, Rendek, K.N, Grotjohann, I, Fromme, P, Kirian, R.A, Beyerlein, K.R, White, T.A, Chapman, H.N, Caffrey, M, Spence, J.C.H, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Serial femtosecond crystallography of G protein-coupled receptors.
Science, 342, 2013
|
|
7WQV
 
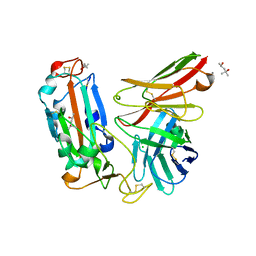 | | Crystal structure of a neutralizing monoclonal antibody (Ab08) in complex with SARS-CoV-2 receptor-binding domain (RBD) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ab08, ... | | Authors: | Zha, J, Meng, L, Zhang, X, Li, D. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Spike-destructing human antibody effectively neutralizes Omicron-included SARS-CoV-2 variants with therapeutic efficacy.
Plos Pathog., 19, 2023
|
|
5XJ7
 
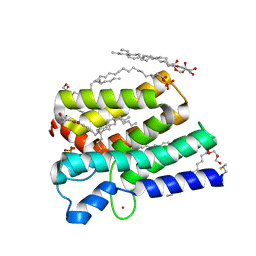 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the acyl phosphate form | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Glycerol-3-phosphate acyltransferase, PHOSPHATE ION, ... | | Authors: | Tang, Y, Li, Z, Li, D. | | Deposit date: | 2017-04-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
5XJ9
 
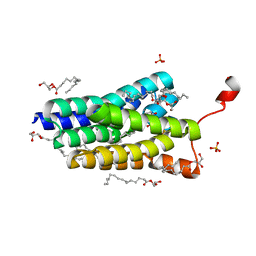 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the orthophosphate form | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Glycerol-3-phosphate acyltransferase, PHOSPHATE ION | | Authors: | Li, Z, Tang, Y, Li, D. | | Deposit date: | 2017-04-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
