6EQI
 
 | | Structure of PINK1 bound to ubiquitin | | Descriptor: | GLYCEROL, Nb696, Serine/threonine-protein kinase PINK1, ... | | Authors: | Schubert, A.F, Gladkova, C, Pardon, E, Wagstaff, J.L, Freund, S.M.V, Steyaert, J, Maslen, S, Komander, D. | | Deposit date: | 2017-10-13 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of PINK1 in complex with its substrate ubiquitin.
Nature, 552, 2017
|
|
4CPH
 
 | | trans-divalent streptavidin with love-hate ligand 4 | | Descriptor: | 5-[(3aS,4S,6aR)-2-oxo-hexahydro-1H-thieno[3,4- d]imidazolidin-4-yl]-N'-{2,6-bis[4-(morpholine-4- sulfonyl)phenyl]phenyl}pentanehydrazide, STREPTAVIDIN | | Authors: | Fairhead, M, Shen, D, Chan, L.K.M, Lowe, E.D, Donohoe, T.J, Howarth, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Love-Hate Ligands for High Resolution Analysis of Strain in Ultra-Stable Protein/Small Molecule Interaction.
Bioorg.Med.Chem., 22, 2014
|
|
6E9D
 
 | | Sub-2 Angstrom Ewald Curvature-Corrected Single-Particle Cryo-EM Reconstruction of AAV-2 L336C | | Descriptor: | Capsid protein VP1 | | Authors: | Tan, Y.Z, Aiyer, S, Mietzsch, M, Hull, J.A, McKenna, R, Baker, T.S, Agbandje-McKenna, M, Lyumkis, D. | | Deposit date: | 2018-07-31 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (1.86 Å) | | Cite: | Sub-2 angstrom Ewald curvature corrected structure of an AAV2 capsid variant.
Nat Commun, 9, 2018
|
|
4CIL
 
 | |
6PNY
 
 | | X-ray Structure of Flpp3 | | Descriptor: | Flpp3 | | Authors: | Zook, J.D, Shekhar, M, Hansen, D.T, Conrad, C, Grant, T.D, Gupta, C, White, T, Barty, A, Basu, S, Zhao, Y, Zatsepin, N.A, Ishchenko, A, Batyuk, A, Gati, C, Li, C, Galli, L, Coe, J, Hunter, M, Liang, M, Weierstall, U, Nelson, G, James, D, Stauch, B, Craciunescu, F, Thifault, D, Liu, W, Cherezov, V, Singharoy, A, Fromme, P. | | Deposit date: | 2019-07-03 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | XFEL and NMR Structures of Francisella Lipoprotein Reveal Conformational Space of Drug Target against Tularemia.
Structure, 28, 2020
|
|
6EOF
 
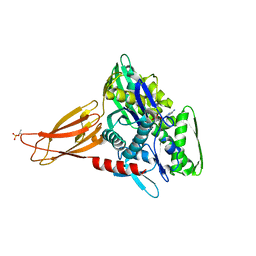 | | Crystal structure of AMPylated GRP78 in ADP state | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Yan, Y, Preissler, S, Read, R.J, Ron, D. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation.
Elife, 6, 2017
|
|
6EPE
 
 | | Substrate processing state 26S proteasome (SPS2) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
6E9X
 
 | | DHF91 filament | | Descriptor: | DHF91 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
4CNZ
 
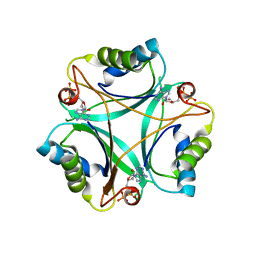 | |
4COM
 
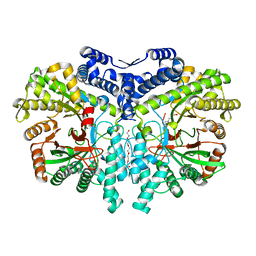 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima with MES in the active site | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, PENTAETHYLENE GLYCOL, ... | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-29 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
4CUE
 
 | | Human Notch1 EGF domains 11-13 mutant T466V | | Descriptor: | CALCIUM ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1 | | Authors: | Taylor, P, Takeuchi, H, Sheppard, D, Chillakuri, C, Lea, S.M, Haltiwanger, R.S, Handford, P.A. | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fringe-Mediated Extension of O-Linked Fucose in the Ligand-Binding Region of Notch1 Increases Binding to Mammalian Notch Ligands.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CO3
 
 | |
4CON
 
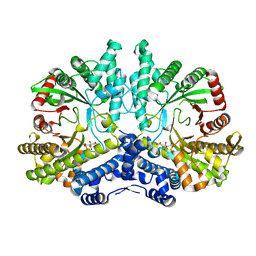 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima with citrate in the active site | | Descriptor: | ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, CITRIC ACID | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-29 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
6EEX
 
 | | L-GSTSTA from degenerate octameric repeats in InaZ, residues 707-712 | | Descriptor: | L-GSTSTA from ice nucleaction protein, inaZ | | Authors: | Zee, C, Glynn, C, Gallagher-Jones, M, Miao, J, Santiago, C.G, Cascio, D, Gonen, T, Sawaya, M.R, Rodriguez, J.A. | | Deposit date: | 2018-08-15 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Homochiral and racemic MicroED structures of a peptide repeat from the ice-nucleation protein InaZ.
IUCrJ, 6, 2019
|
|
6E6E
 
 | | DGY-06-116, a novel and selective covalent inhibitor of SRC kinase | | Descriptor: | N-(2-chloro-6-methylphenyl)-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-4-{[2-(propanoylamino)phenyl]amino}pyrimidine-5-carboxamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Gurbani, D, Bera, A, Westover, K. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Characterization of a Covalent Inhibitor of Src Kinase.
Front Mol Biosci, 7, 2020
|
|
4D5H
 
 | | Focal Adhesion Kinase catalytic domain | | Descriptor: | 6-methyl-5-{[3-(trifluoromethyl)phenyl]amino}-1,2,4-triazin-3(4H)-one, FOCAL ADHESION KINASE 1, SULFATE ION | | Authors: | Le Coq, J, Lin, A, Lietha, D. | | Deposit date: | 2014-11-05 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Allosteric Regulation of Focal Adhesion Kinase by Pip2 and ATP.
Biophys.J., 108, 2015
|
|
4D0F
 
 | | Human Notch1 EGF domains 11-13 mutant T466A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1 | | Authors: | Taylor, P, Takeuchi, H, Sheppard, D, Chillakuri, C, Lea, S.M, Haltiwanger, R.S, Handford, P.A. | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fringe-Mediated Extension of O-Linked Fucose in the Ligand-Binding Region of Notch1 Increases Binding to Mammalian Notch Ligands.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CO7
 
 | | Crystal structure of human GATE-16 | | Descriptor: | GAMMA-AMINOBUTYRIC ACID RECEPTOR-ASSOCIATED PROTEIN-LIKE 2 | | Authors: | Weiergraeber, O.H, Ma, P, Willbold, D. | | Deposit date: | 2014-01-27 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Polymorphism in Autophagy-Related Protein Gate-16.
Biochemistry, 54, 2015
|
|
4D7T
 
 | | Structure of the SthK Carboxy-Terminal Region in complex with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, STHK_CNBD_CAMP | | Authors: | Kesters, D, Brams, M, Nys, M, Wijckmans, E, Spurny, R, Voets, T, Tytgat, J, Ulens, C. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.582 Å) | | Cite: | Structure of the SthK Carboxy-Terminal Region Reveals a Gating Mechanism for Cyclic Nucleotide-Modulated Ion Channels.
Plos One, 10, 2015
|
|
4CPF
 
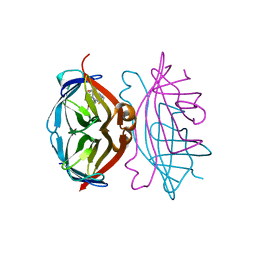 | | Wild-type streptavidin in complex with love-hate ligand 3 (LH3) | | Descriptor: | STREPTAVIDIN, methyl 4-(2-{5-[(3aS,4S,6aR)-2-oxo-hexahydro-1H- thieno[3,4-d]imidazolidin-4-yl]pentanehydrazido}-3- [4-(methoxycarbonyl)phenyl]phenyl)benzoate | | Authors: | Fairhead, M, Shen, D, Chan, L.K.M, Lowe, E.D, Donohoe, T.J, Howarth, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Love-Hate Ligands for High Resolution Analysis of Strain in Ultra-Stable Protein/Small Molecule Interaction.
Bioorg.Med.Chem., 22, 2014
|
|
4D5K
 
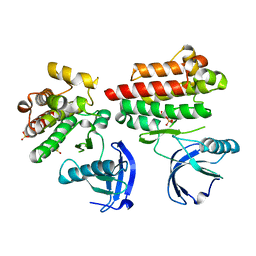 | | Focal Adhesion Kinase catalytic domain | | Descriptor: | DIMETHYL SULFOXIDE, FOCAL ADHESION KINASE, SULFATE ION | | Authors: | Le Coq, J, Lin, A, Lietha, D. | | Deposit date: | 2014-11-05 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Allosteric Regulation of Focal Adhesion Kinase by Pip2 and ATP.
Biophys.J., 108, 2015
|
|
6EBL
 
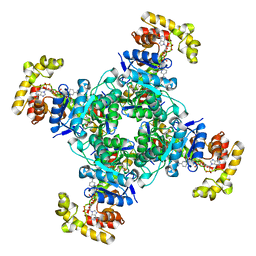 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs, cytosolic domain | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera, Voltage-gated potassium channel subunit beta-2 | | Authors: | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | Deposit date: | 2018-08-06 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
4D7L
 
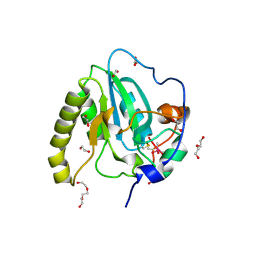 | | Methionine sulfoxide reductase A of Corynebacterium diphtheriae | | Descriptor: | CACODYLATE ION, PEPTIDE METHIONINE SULFOXIDE REDUCTASE MSRA, SULFATE ION, ... | | Authors: | Van Molle, I, Tossounian, M.A, Pedre, B, Wahni, K, Vertommen, D, Messens, J. | | Deposit date: | 2014-11-25 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Corynebacterium Diphtheriae Methionine Sulfoxide Reductase a Exploits a Unique Mycothiol Redox Relay Mechanism.
J.Biol.Chem., 290, 2015
|
|
6EHY
 
 | |
6ERE
 
 | | Crystal structure of a computationally designed colicin endonuclease and immunity pair colEdes3/Imdes3 | | Descriptor: | Immunity, PHOSPHATE ION, colicin | | Authors: | Netzer, R, Listov, D, Dym, O, Albeck, S, Knop, O, Fleishman, S.J. | | Deposit date: | 2017-10-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Ultrahigh specificity in a network of computationally designed protein-interaction pairs.
Nat Commun, 9, 2018
|
|
