9EEI
 
 | |
9EET
 
 | |
9IVP
 
 | | 24-mer DARPin-apoferritin scaffold in complex with the maltose binding protein | | Descriptor: | DARPin,Ferritin heavy chain, N-terminally processed, Maltodextrin-binding protein | | Authors: | Lu, X, Yan, M, Zhang, H.M, Hao, Q. | | Deposit date: | 2024-07-24 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A large, general and modular DARPin-apoferritin scaffold enables the visualization of small proteins by cryo-EM.
Iucrj, 12, 2025
|
|
7XF5
 
 | | Full length human CLC-2 channel in apo state | | Descriptor: | Chloride channel protein 2 | | Authors: | Wang, L. | | Deposit date: | 2022-04-01 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of ClC-2 chloride channel reveal the blocking mechanism of its specific inhibitor AK-42
Nat Commun, 14, 2023
|
|
7XJA
 
 | | TMD masked refine map of human ClC-2 | | Descriptor: | Chloride channel protein 2 | | Authors: | Wang, L. | | Deposit date: | 2022-04-15 | | Release date: | 2023-05-17 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of ClC-2 chloride channel reveal the blocking mechanism of its specific inhibitor AK-42
Nat Commun, 14, 2023
|
|
6X5Y
 
 | | IDO1 in complex with compound 4 | | Descriptor: | 4-fluoro-N-{1-[5-(2-methylpyrimidin-4-yl)-5,6,7,8-tetrahydro-1,5-naphthyridin-2-yl]cyclopropyl}benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A, Lammens, A. | | Deposit date: | 2020-05-27 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Utilization of MetID and Structural Data to Guide Placement of Spiro and Fused Cyclopropyl Groups for the Synthesis of Low Dose IDO1 Inhibitors
To Be Published
|
|
6J1U
 
 | | influenza virus nucleoprotein with a specific inhibitor | | Descriptor: | Nucleoprotein, ~{N}-[4-[(4-~{tert}-butylphenyl)carbonylamino]phenyl]-2,3-dihydro-1,4-benzodioxine-6-carboxamide | | Authors: | Pang, B, Zhang, W.Z, Zhang, H.M, Hao, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a Novel Specific Inhibitor Targeting Influenza A Virus Nucleoprotein with Pleiotropic Inhibitory Effects on Various Steps of the Viral Life Cycle.
J.Virol., 95, 2021
|
|
7XED
 
 | | Crystal Structure of OsCIE1-Ubox and OsUBC8 complex | | Descriptor: | U-box domain-containing protein 12, UBC core domain-containing protein | | Authors: | Zhang, Y, Yu, C.Z. | | Deposit date: | 2022-03-30 | | Release date: | 2023-10-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Release of a ubiquitin brake activates OsCERK1-triggered immunity in rice.
Nature, 629, 2024
|
|
1BA2
 
 | |
7JZW
 
 | | Cryo-EM structure of CRISPR-Cas surveillance complex with AcrIF4 | | Descriptor: | CRISPR repeat sequence, CRISPR type I-F/YPEST-associated protein Csy1, CRISPR type I-F/YPEST-associated protein Csy2, ... | | Authors: | Chang, L, Li, Z, Gabel, C. | | Deposit date: | 2020-09-02 | | Release date: | 2020-12-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for inhibition of the type I-F CRISPR-Cas surveillance complex by AcrIF4, AcrIF7 and AcrIF14.
Nucleic Acids Res., 49, 2021
|
|
7JZX
 
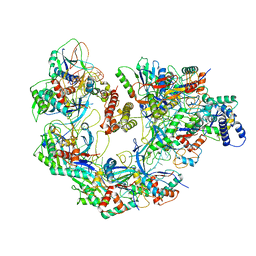 | | Cryo-EM structure of CRISPR-Cas surveillance complex with AcrIF7 | | Descriptor: | AcrF7, CRISPR type I-F/YPEST-associated protein Csy3, CRISPR-associated endonuclease Cas6/Csy4, ... | | Authors: | Chang, L, Li, Z, Gabel, C. | | Deposit date: | 2020-09-02 | | Release date: | 2020-12-30 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for inhibition of the type I-F CRISPR-Cas surveillance complex by AcrIF4, AcrIF7 and AcrIF14.
Nucleic Acids Res., 49, 2021
|
|
7JZZ
 
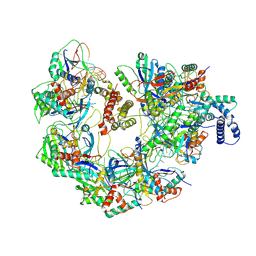 | | Cryo-EM structure of CRISPR-Cas surveillance complex with AcrIF14 | | Descriptor: | AcrF14, CRISPR type I-F/YPEST-associated protein Csy3, CRISPR-associated protein Csy1, ... | | Authors: | Chang, L, Li, Z, Gabel, C. | | Deposit date: | 2020-09-02 | | Release date: | 2020-12-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for inhibition of the type I-F CRISPR-Cas surveillance complex by AcrIF4, AcrIF7 and AcrIF14.
Nucleic Acids Res., 49, 2021
|
|
8STB
 
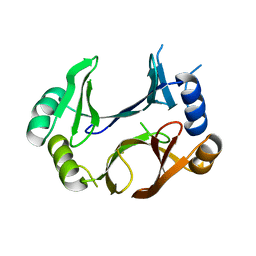 | | The structure of abxF, an enzyme catalyzing the formation of the chiral spiroketal of an anthrabenzoxocinone antibiotic, (-)-ABX | | Descriptor: | GLYCEROL, Glyoxalase, SULFATE ION, ... | | Authors: | Luo, Z, Jia, X, Yan, X, Qu, X, Kobe, B. | | Deposit date: | 2023-05-09 | | Release date: | 2024-05-22 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | An enzymatic dual-oxa Diels-Alder reaction constructs the oxygen-bridged tricyclic acetal unit of (-)-anthrabenzoxocinone.
Nat.Chem., 2025
|
|
9LNH
 
 | | Crystal structure of Peroxiredoxin I in complex with compound LC-PDin20 | | Descriptor: | (2~{S})-2-[[(2~{R},4~{a}~{S},6~{a}~{R},6~{a}~{S},14~{a}~{S},14~{b}~{R})-2,4~{a},6~{a},6~{a},9,14~{a}-hexamethyl-10-oxidanyl-11-oxidanylidene-1,3,4,5,6,13,14,14~{b}-octahydropicen-2-yl]carbamoylamino]butanoic acid, Peroxiredoxin-1 | | Authors: | Wang, Z, Luo, C. | | Deposit date: | 2025-01-21 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Rapid Discovery of Celastrol Derivatives as Potent and Selective PRDX1 Inhibitors via Microplate-Based Parallel Compound Library and In Situ Screening.
J.Med.Chem., 2025
|
|
8K14
 
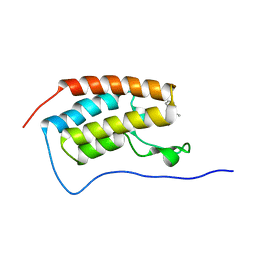 | | X-ray crystal structure of 18a in BRD4(1) | | Descriptor: | 4-[8-methoxy-2-methyl-1-(1-phenylethyl)imidazo[4,5-c]quinolin-7-yl]-3,5-dimethyl-1,2-oxazole, Bromodomain-containing protein 4 | | Authors: | Xu, H, Shen, H, Zhang, Y, Xu, Y, Li, R. | | Deposit date: | 2023-07-10 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Discovery of (R)-4-(8-methoxy-2-methyl-1-(1-phenylethy)-1H-imidazo[4,5-c]quinnolin-7-yl)-3,5-dimethylisoxazole as a potent and selective BET inhibitor for treatment of acute myeloid leukemia (AML) guided by FEP calculation.
Eur.J.Med.Chem., 263, 2024
|
|
8VLR
 
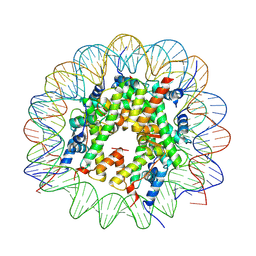 | | Cryo-EM structure of native H2AK119bu nucleosome at 2.6 | | Descriptor: | DNA (136-MER), Histone H2A type 1-B/E, Histone H2B type 1-A, ... | | Authors: | Wang, Y, Zhang, K. | | Deposit date: | 2024-01-12 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Role of histone variants H2BC1 and H2AZ.2 in H2AK119ub nucleosome organization and Polycomb gene silencing
To Be Published
|
|
7XF3
 
 | | The structure of HLA-B*1501/BM58-66AF9 | | Descriptor: | 9-mer peptide from Matrix protein 1, Beta-2-microglobulin, MHC class I antigen | | Authors: | Zhao, Y.Z, Xiao, W.L, Wu, Y.N, Fan, W.F, Yue, C, Zhang, Q.X, Zhang, D.N, Yuan, X.J, Yao, S.J, Liu, S, Li, M, Wang, P.Y, Zhang, H.J, Zhang, J, Zhao, M, Zheng, X.Q, Liu, W.J, Gao, G.F, Liu, W.L. | | Deposit date: | 2022-03-31 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Parallel T Cell Immunogenic Regions in Influenza B and A Viruses with Distinct Nuclear Export Signal Functions: The Balance between Viral Life Cycle and Immune Escape.
J Immunol., 210, 2023
|
|
8OWO
 
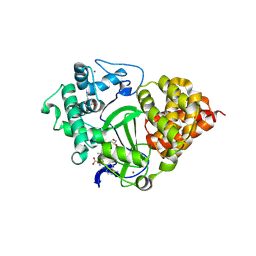 | | SMYD3 in complex with fragment FL01507 | | Descriptor: | 3-oxidanylbenzenecarbonitrile, GLYCEROL, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Lund, B.A, Cederfelt, D, Dobritzsch, D. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of fragments targeting SMYD3 using highly sensitive kinetic and multiplexed biosensor-based screening.
Rsc Med Chem, 15, 2024
|
|
9KRD
 
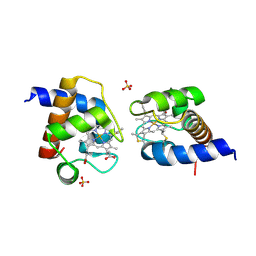 | | CRYSTAL STRUCTURE OF REDUCED CYTOCHROME C6 FROM SYNECHOCOCCUS ELONGATUS PCC 7942 | | Descriptor: | Cytochrome c6, HEME C, SULFATE ION | | Authors: | Zhang, B.T, Liu, S.W, Xu, Y.C, Sheng, W, Gong, Y, Cao, P. | | Deposit date: | 2024-11-27 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A High-Resolution Crystallographic Study of Cytochrome c6: Structural Basis for Electron Transfer in Cyanobacterial Photosynthesis.
Int J Mol Sci, 26, 2025
|
|
9KRC
 
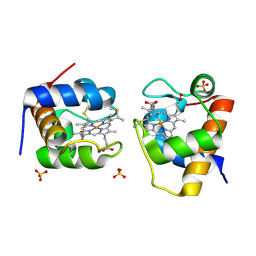 | | CRYSTAL STRUCTURE OF OXIDIIZED CYTOCHROME C6 FROM SYNECHOCOCCUS ELONGATUS PCC 7942 | | Descriptor: | Cytochrome c6, HEME C, SULFATE ION | | Authors: | Zhang, B.T, Liu, S.W, Xu, Y.C, Sheng, W, Gong, Y, Cao, P. | | Deposit date: | 2024-11-27 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A High-Resolution Crystallographic Study of Cytochrome c6: Structural Basis for Electron Transfer in Cyanobacterial Photosynthesis.
Int J Mol Sci, 26, 2025
|
|
9KRR
 
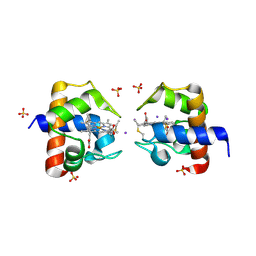 | | CRYSTAL STRUCTURE OF REDUCED CYTOCHROME C6 FROM Synechocystis PCC 6803 | | Descriptor: | Cytochrome c6, HEME C, IODIDE ION, ... | | Authors: | Zhang, B.T, Liu, S.W, Xu, Y.C, Sheng, W, Gong, Y, Cao, P. | | Deposit date: | 2024-11-28 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A High-Resolution Crystallographic Study of Cytochrome c6: Structural Basis for Electron Transfer in Cyanobacterial Photosynthesis.
Int J Mol Sci, 26, 2025
|
|
8XD7
 
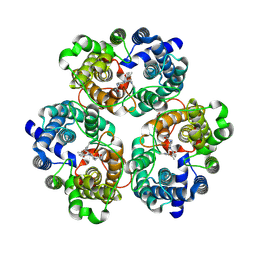 | |
8XDB
 
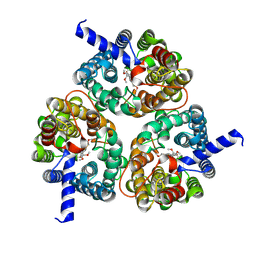 | | Cryo-EM structure of human urea transporter A2. | | Descriptor: | (5E)-2-azanylidene-5-[(2,3-dimethoxyphenyl)methylidene]-1,3-thiazolidin-4-one, Urea transporter 2 | | Authors: | Huang, S, Liu, L, Sun, J. | | Deposit date: | 2023-12-10 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the mechanisms of urea permeation and distinct inhibition modes of urea transporters.
Nat Commun, 15, 2024
|
|
8XDC
 
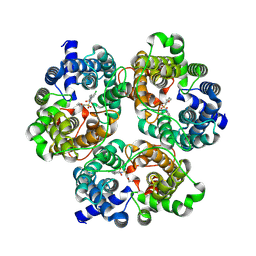 | | Cryo-EM structure of human urea transporter A2. | | Descriptor: | N-[3-[1,1-bis(oxidanylidene)-1,2-thiazolidin-2-yl]-4-chloranyl-phenyl]-2-methoxy-5-methyl-benzenesulfonamide, Urea transporter 2 | | Authors: | Huang, S, Liu, L, Sun, J. | | Deposit date: | 2023-12-10 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the mechanisms of urea permeation and distinct inhibition modes of urea transporters.
Nat Commun, 15, 2024
|
|
8XDI
 
 | | Cryo-EM structure of zebrafish urea transporter. | | Descriptor: | N-(4-acetamidophenyl)-5-ethanoyl-furan-2-carboxamide, Urea transporter | | Authors: | Huang, S, Liu, L, Sun, J. | | Deposit date: | 2023-12-11 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the mechanisms of urea permeation and distinct inhibition modes of urea transporters.
Nat Commun, 15, 2024
|
|
