2MUR
 
 | |
2ZZ8
 
 | |
3AIB
 
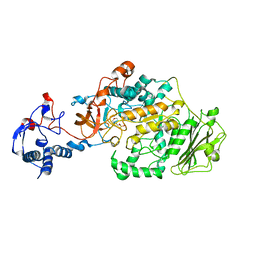 | | Crystal Structure of Glucansucrase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Glucosyltransferase-SI, ... | | Authors: | Ito, K, Ito, S, Shimamura, T, Iwata, S. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans.
J.Mol.Biol., 408, 2011
|
|
3BEJ
 
 | | Structure of human FXR in complex with MFA-1 and co-activator peptide | | Descriptor: | (8alpha,10alpha,13alpha,17beta)-17-[(4-hydroxyphenyl)carbonyl]androsta-3,5-diene-3-carboxylic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Soisson, S.M, Parthasarathy, G, Becker, J.W. | | Deposit date: | 2007-11-19 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a potent synthetic FXR agonist with an unexpected mode of binding and activation.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3AQP
 
 | | Crystal structure of SecDF, a translocon-associated membrane protein, from Thermus thrmophilus | | Descriptor: | Probable SecDF protein-export membrane protein | | Authors: | Tsukazaki, T, Mori, H, Echizen, Y, Ishitani, R, Fukai, S, Tanaka, T, Perederina, A, Vassylyev, D.G, Kohno, T, Ito, K, Nureki, O. | | Deposit date: | 2010-11-16 | | Release date: | 2011-05-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and function of a membrane component SecDF that enhances protein export
Nature, 474, 2011
|
|
3AQO
 
 | |
3BPN
 
 | | Crystal structure of the IL4-IL4R-IL13Ra ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-13 receptor alpha-1 chain, Interleukin-4, ... | | Authors: | Garcia, K.C. | | Deposit date: | 2007-12-18 | | Release date: | 2008-02-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Molecular and Structural Basis of Cytokine Receptor Pleiotropy in the Interleukin-4/13 System.
Cell(Cambridge,Mass.), 132, 2008
|
|
3BPL
 
 | | Crystal structure of the IL4-IL4R-Common Gamma ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Garcia, K.C. | | Deposit date: | 2007-12-18 | | Release date: | 2008-02-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Molecular and Structural Basis of Cytokine Receptor Pleiotropy in the Interleukin-4/13 System.
Cell(Cambridge,Mass.), 132, 2008
|
|
3BPO
 
 | | Crystal structure of the IL13-IL4R-IL13Ra ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin 13, ... | | Authors: | Garcia, K.C. | | Deposit date: | 2007-12-18 | | Release date: | 2008-02-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular and Structural Basis of Cytokine Receptor Pleiotropy in the Interleukin-4/13 System.
Cell(Cambridge,Mass.), 132, 2008
|
|
2Q11
 
 | | Structure of BACE complexed to compound 1 | | Descriptor: | 3-(2-AMINO-6-BENZOYLQUINAZOLIN-3(4H)-YL)-N-CYCLOHEXYL-N-METHYLPROPANAMIDE, Beta-secretase 1 | | Authors: | Sharff, A.J. | | Deposit date: | 2007-05-23 | | Release date: | 2007-08-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-Amino-3,4-dihydroquinazolines as inhibitors of BACE-1 (beta-Site APP cleaving enzyme): Use of structure based design to convert a micromolar hit into a nanomolar lead.
J.Med.Chem., 50, 2007
|
|
2Q15
 
 | | Structure of BACE complexed to compound 3a | | Descriptor: | (4S)-4-(2-AMINO-6-PHENOXYQUINAZOLIN-3(4H)-YL)-N,4-DICYCLOHEXYL-N-METHYLBUTANAMIDE, Beta-secretase 1 | | Authors: | Sharff, A.J. | | Deposit date: | 2007-05-23 | | Release date: | 2007-08-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-Amino-3,4-dihydroquinazolines as inhibitors of BACE-1 (beta-Site APP cleaving enzyme): Use of structure based design to convert a micromolar hit into a nanomolar lead.
J.Med.Chem., 50, 2007
|
|
3AIE
 
 | | Crystal Structure of glucansucrase from Streptococcus mutans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Glucosyltransferase-SI | | Authors: | Ito, K, Ito, S, Shimamura, T, Iwata, S. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans.
J.Mol.Biol., 408, 2011
|
|
3AIC
 
 | | Crystal Structure of Glucansucrase from Streptococcus mutans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ito, K, Ito, S, Shimamura, T, Iwata, S. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans.
J.Mol.Biol., 408, 2011
|
|
2OMW
 
 | | Crystal structure of InlA S192N Y369S/mEC1 complex | | Descriptor: | CHLORIDE ION, Epithelial-cadherin, Internalin-A | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Extending the host range of Listeria monocytogenes by rational protein design.
Cell(Cambridge,Mass.), 129, 2007
|
|
3CR5
 
 | | X-ray structure of bovine Pnt-Zn(2+),Ca(2+)-S100B | | Descriptor: | 1,5-BIS(4-AMIDINOPHENOXY)PENTANE, CALCIUM ION, Protein S100-B, ... | | Authors: | Charpentier, T.H. | | Deposit date: | 2008-04-04 | | Release date: | 2008-06-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Divalent metal ion complexes of S100B in the absence and presence of pentamidine.
J.Mol.Biol., 382, 2008
|
|
3CTQ
 
 | | Structure of MAP kinase p38 in complex with a 1-o-tolyl-1,2,3-triazole-4-carboxamide | | Descriptor: | Mitogen-activated protein kinase 14, N-benzyl-1-[5-({5-tert-butyl-2-methoxy-3-[(methylsulfonyl)amino]phenyl}carbamoyl)-2-methylphenyl]-1H-1,2,3-triazole-4-carboxamide | | Authors: | Qian, K. | | Deposit date: | 2008-04-14 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design and subsequent optimization of 2-tolyl-(1,2,3-triazol-1-yl-4-carboxamide) inhibitors of p38 MAP kinase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3D14
 
 | | Crystal structure of mouse Aurora A (Asn186->Gly, Lys240->Arg, Met302->Leu) in complex with 1-{5-[2-(thieno[3,2-d]pyrimidin-4-ylamino)-ethyl]- thiazol-2-yl}-3-(3-trifluoromethyl-phenyl)-urea | | Descriptor: | 1-{5-[2-(thieno[3,2-d]pyrimidin-4-ylamino)ethyl]-1,3-thiazol-2-yl}-3-[3-(trifluoromethyl)phenyl]urea, serine/threonine kinase 6 | | Authors: | Elling, R.A, Baskaran, S, Allen, D.A, Oslob, J.D, Romanowski, M.J. | | Deposit date: | 2008-05-04 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a potent and selective aurora kinase inhibitor.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2OIN
 
 | | crystal structure of HCV NS3-4A R155K mutant | | Descriptor: | NS4A peptide, Polyprotein, ZINC ION | | Authors: | Wei, Y. | | Deposit date: | 2007-01-11 | | Release date: | 2007-06-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phenotypic and structural analyses of hepatitis C virus NS3 protease Arg155 variants: sensitivity to telaprevir (VX-950) and interferon alpha.
J.Biol.Chem., 282, 2007
|
|
2OMV
 
 | | Crystal structure of InlA S192N Y369S/hEC1 complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epithelial-cadherin, ... | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Extending the host range of Listeria monocytogenes by rational protein design.
Cell(Cambridge,Mass.), 129, 2007
|
|
2OMY
 
 | | Crystal structure of InlA S192N/hEC1 complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epithelial-cadherin, ... | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extending the host range of Listeria monocytogenes by rational protein design.
Cell(Cambridge,Mass.), 129, 2007
|
|
2QIQ
 
 | |
3CR2
 
 | | X-ray structure of bovine Zn(2+),Ca(2+)-S100B | | Descriptor: | CALCIUM ION, Protein S100-B, ZINC ION | | Authors: | Charpentier, T.H. | | Deposit date: | 2008-04-04 | | Release date: | 2008-06-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Divalent metal ion complexes of S100B in the absence and presence of pentamidine.
J.Mol.Biol., 382, 2008
|
|
3D15
 
 | | Crystal structure of mouse Aurora A (Asn186->Gly, Lys240->Arg, Met302->Leu) in complex with 1-(3-chloro-phenyl)-3-{5-[2-(thieno[3,2-d]pyrimidin-4-ylamino)- ethyl]-thiazol-2-yl}-urea [SNS-314] | | Descriptor: | 1-(3-chlorophenyl)-3-{5-[2-(thieno[3,2-d]pyrimidin-4-ylamino)ethyl]-1,3-thiazol-2-yl}urea, serine/threonine kinase 6 | | Authors: | Elling, R.A, Bui, M, Allen, D.A, Oslob, J.D, Romanowski, M.J. | | Deposit date: | 2008-05-04 | | Release date: | 2009-05-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a potent and selective aurora kinase inhibitor.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CR4
 
 | | X-ray structure of bovine Pnt,Ca(2+)-S100B | | Descriptor: | 1,5-BIS(4-AMIDINOPHENOXY)PENTANE, CALCIUM ION, Protein S100-B | | Authors: | Charpentier, T.H. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Divalent metal ion complexes of S100B in the absence and presence of pentamidine.
J.Mol.Biol., 382, 2008
|
|
2R2L
 
 | | Structure of Farnesyl Protein Transferase bound to PB-93 | | Descriptor: | FARNESYL DIPHOSPHATE, Farnesyltransferase subunit alpha, Farnesyltransferase subunit beta, ... | | Authors: | Strickland, C.O, Voorhis, W. | | Deposit date: | 2007-08-27 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Efficacy, pharmacokinetics, and metabolism of tetrahydroquinoline inhibitors of Plasmodium falciparum protein farnesyltransferase.
Antimicrob.Agents Chemother., 51, 2007
|
|
