1ZGX
 
 | |
1TO9
 
 | |
1TOO
 
 | | Interleukin 1B Mutant F146W | | Descriptor: | Interleukin-1 beta | | Authors: | Adamek, D.H, Guerrero, L, Caspar, D.L. | | Deposit date: | 2004-06-14 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and energetic consequences of mutations in a solvated hydrophobic cavity.
J.Mol.Biol., 346, 2005
|
|
1TP0
 
 | |
1ZAE
 
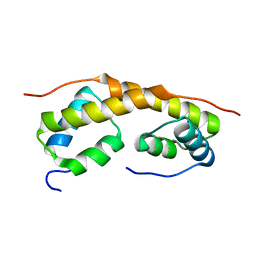 | | Solution structure of the functional domain of phi29 replication organizer p16.7c | | Descriptor: | Early protein GP16.7 | | Authors: | Asensio, J.L, Albert, A, Munoz-Espin, D, Gonzalez, C, Hermoso, J, Villar, L, Jimenez-Barbero, J, Salas, M, Meijer, W.J.J. | | Deposit date: | 2005-04-06 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the functional domain of phi29 replication organizer: insights into oligomerization and dna binding
J.Biol.Chem., 280, 2005
|
|
1TT7
 
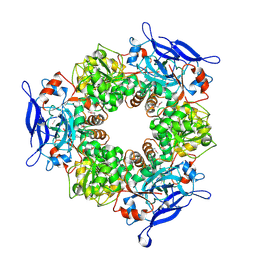 | |
5EYV
 
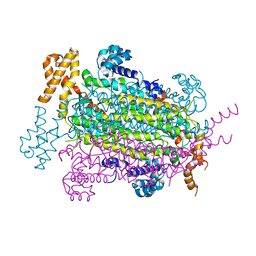 | | Crystal Structure of Adenylosuccinate lyase from Schistosoma mansoni in APO form. | | Descriptor: | Adenylosuccinate lyase | | Authors: | Romanello, L, Torini, J.R, Bird, L.E, Nettleship, J.E, Owens, R.J, Reddivari, Y, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2015-11-25 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural and kinetic analysis of Schistosoma mansoni Adenylosuccinate Lyase (SmADSL).
Mol. Biochem. Parasitol., 214, 2017
|
|
1ZNU
 
 | | Structure of cyclotide Kalata B1 in DPC micelles solution | | Descriptor: | Kalata B1 | | Authors: | Shenkarev, Z.O, Nadezhdin, K.D, Sobol, V.A, Sobol, A.G, Skjeldal, L, Arseniev, A.S. | | Deposit date: | 2005-05-12 | | Release date: | 2006-04-25 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Conformation and mode of membrane interaction in cyclotides. Spatial structure of kalata B1 bound to a dodecylphosphocholine micelle.
Febs J., 273, 2006
|
|
1TVL
 
 | |
1ZPC
 
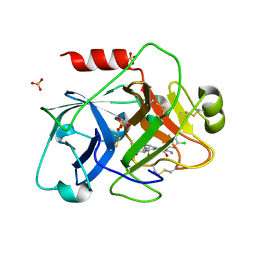 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with 2-[2-(3-Chloro-phenyl)-2-hydroxy-acetylamino]-N-[4-guanidino-1-(thiazole-2-carbonyl)-butyl]-3-methyl-butyramide | | Descriptor: | 2-[2-(3-CHLORO-PHENYL)-2-HYDROXY-ACETYLAMINO]-N-[4-GUANIDINO-1-(THIAZOLE-2-CARBONYL)-BUTYL]-3-METHYL-BUTYRAMIDE, Coagulation factor XI, SULFATE ION | | Authors: | Deng, H, Bannister, T.D, Jin, L, Nagafuji, P, Celatka, C.A, Lin, J, Lazarova, T.I, Rynkiewicz, M.J, Quinn, J, Bibbins, F, Pandey, P, Gorga, J, Babine, R.E, Meyers, H.V, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2005-05-16 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis, SAR exploration, and X-ray crystal structures of factor XIa inhibitors containing an alpha-ketothiazole arginine
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1YFK
 
 | | Crystal structure of human B type phosphoglycerate mutase | | Descriptor: | CHLORIDE ION, CITRIC ACID, Phosphoglycerate mutase 1 | | Authors: | Wang, Y, Wei, Z, Liu, L, Gong, W. | | Deposit date: | 2005-01-02 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human B-type phosphoglycerate mutase bound with citrate.
Biochem.Biophys.Res.Commun., 331, 2005
|
|
5EWV
 
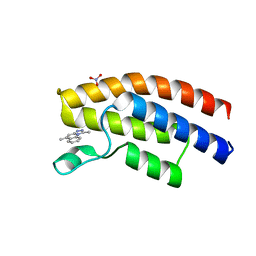 | | Crystal structure of the human BRPF1 bromodomain in complex with SEED20 | | Descriptor: | 1,5-dimethyl-[1,2,4]triazolo[4,3-a]quinoline, NITRATE ION, Peregrin | | Authors: | Zhu, J, Wiedmer, L, Caflisch, A. | | Deposit date: | 2015-11-21 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of the human BRPF1 bromodomain in complex with SEED20
To Be Published
|
|
1TO3
 
 | |
1TOB
 
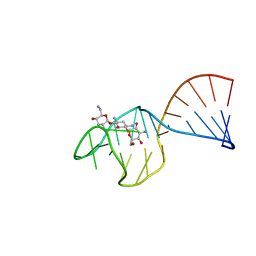 | | SACCHARIDE-RNA RECOGNITION IN AN AMINOGLYCOSIDE ANTIBIOTIC-RNA APTAMER COMPLEX, NMR, 7 STRUCTURES | | Descriptor: | 1,3-DIAMINO-5,6-DIHYDROXYCYCLOHEXANE, 2,6-diammonio-2,3,6-trideoxy-alpha-D-glucopyranose, 3-ammonio-3-deoxy-alpha-D-glucopyranose, ... | | Authors: | Jiang, L, Suri, A.K, Fiala, R, Patel, D.J. | | Deposit date: | 1996-12-12 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Saccharide-RNA recognition in an aminoglycoside antibiotic-RNA aptamer complex.
Chem.Biol., 4, 1997
|
|
1TQE
 
 | | Mechanism of recruitment of class II histone deacetylases by myocyte enhancer factor-2 | | Descriptor: | Histone deacetylase 9, MEF2 binding site of nur77 promoter, Myocyte-specific enhancer factor 2B | | Authors: | Chen, L, Han, A, He, J, Wu, Y, Liu, J.O. | | Deposit date: | 2004-06-17 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of Recruitment of Class II Histone Deacetylases by Myocyte Enhancer Factor-2.
J.Mol.Biol., 345, 2005
|
|
1TP7
 
 | | Crystal Structure of the RNA-dependent RNA Polymerase from Human Rhinovirus 16 | | Descriptor: | 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, Genome polyprotein, SULFATE ION | | Authors: | Appleby, T.C, Luecke, H, Shim, J.H, Wu, J.Z, Cheney, I.W, Zhong, W, Vogeley, L, Hong, Z, Yao, N. | | Deposit date: | 2004-06-15 | | Release date: | 2005-06-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of complete rhinovirus RNA polymerase suggests front loading of protein primer.
J.Virol., 79, 2005
|
|
1TQF
 
 | | Crystal structure of human Beta secretase complexed with inhibitor | | Descriptor: | 3-{2-[(5-AMINOPENTYL)AMINO]-2-OXOETHOXY}-5-({[1-(4-FLUOROPHENYL)ETHYL]AMINO}CARBONYL)PHENYL PHENYLMETHANESULFONATE, Beta-secretase 1 | | Authors: | Munshi, S, Chen, Z, Kuo, L. | | Deposit date: | 2004-06-17 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a small molecule nonpeptide active site beta-secretase inhibitor that displays a nontraditional binding mode for aspartyl proteases.
J.Med.Chem., 47, 2004
|
|
1YIS
 
 | | Structural genomics of Caenorhabditis elegans: adenylosuccinate lyase | | Descriptor: | SULFATE ION, adenylosuccinate lyase | | Authors: | Symersky, J, Schormann, N, Lu, S, Zhang, Y, Karpova, E, Qiu, S, Huang, W, Cao, Z, Zhou, J, Luo, M, Arabshahi, A, McKinstry, A, Luan, C.-H, Luo, D, Johnson, D, An, J, Tsao, J, Delucas, L, Shang, Q, Gray, R, Li, S, Bray, T, Chen, Y.-J, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-01-12 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: adenylosuccinate lyase
To be Published
|
|
1Y2K
 
 | | Catalytic Domain Of Human Phosphodiesterase 4D In Complex With 3,5-dimethyl-1-(3-nitro-phenyl)-1H-pyrazole-4-carboxylic acid ethyl ester | | Descriptor: | 1,2-ETHANEDIOL, 3,5-DIMETHYL-1-(3-NITROPHENYL)-1H-PYRAZOLE-4-CARBOXYLIC ACID ETHYL ESTER, MAGNESIUM ION, ... | | Authors: | Card, G.L, Blasdel, L, England, B.P, Zhang, C, Suzuki, Y, Gillette, S, Fong, D, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-11-22 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A family of phosphodiesterase inhibitors discovered by cocrystallography and scaffold-based drug design
Nat.Biotechnol., 23, 2005
|
|
1TZ7
 
 | | Aquifex aeolicus amylomaltase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-alpha-glucanotransferase | | Authors: | Barends, T.R.M, Korf, H, Kaper, T, van der Maarel, M.J.E.C, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2004-07-09 | | Release date: | 2005-08-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural influences on product specificity in amylomaltase from Aquifex aeolicus
TO BE PUBLISHED
|
|
1Y7W
 
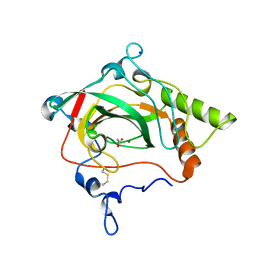 | | Crystal structure of a halotolerant carbonic anhydrase from Dunaliella salina | | Descriptor: | ACETIC ACID, Halotolerant alpha-type carbonic anhydrase (dCA II), SODIUM ION, ... | | Authors: | Premkumar, L, Greenblatt, H.M, Bageshwar, U.K, Savchenko, T, Gokhman, I, Sussman, J.L, Zamir, A, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2004-12-10 | | Release date: | 2005-05-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Three-dimensional structure of a halotolerant algal carbonic anhydrase predicts halotolerance of a mammalian homolog.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6IQ4
 
 | | Nucleosome core particle cross-linked with a hetero-binuclear molecule possessing RAPTA and gold(I) 4-(diphenylphosphino)benzoic acid groups. | | Descriptor: | 4-diphenylphosphanylbenzoic acid, DNA (145-MER), GOLD ION, ... | | Authors: | DeFalco, L, Batchelor, L.K, Adhireksan, Z, Dyson, P.J, Davey, C.A. | | Deposit date: | 2018-11-06 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crosslinking Allosteric Sites on the Nucleosome.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
1YBI
 
 | | Crystal structure of HA33A, a neurotoxin-associated protein from Clostridium botulinum type A | | Descriptor: | non-toxin haemagglutinin HA34 | | Authors: | Arndt, J.W, Gu, J, Jaroszewski, L, Schwarzenbacher, R, Hanson, M, Lebeda, F.J, Stevens, R.C. | | Deposit date: | 2004-12-20 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of the Neurotoxin-associated Protein HA33/A from Clostridium botulinum Suggests a Reoccurring beta-Trefoil Fold in the Progenitor Toxin Complex.
J.Mol.Biol., 346, 2005
|
|
1YBZ
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-1581948-001 | | Descriptor: | UNKNOWN ATOM OR ION, chorismate mutase | | Authors: | Lee, D, Chen, L, Nguyen, D, Dillard, B.D, Tempel, W, Habel, J, Zhou, W, Chang, S.-H, Kelley, L.-L.C, Liu, Z.-J, Lin, D, Zhang, H, Praissman, J, Bridger, S, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.-S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Conserved hypothetical protein from Pyrococcus furiosus Pfu-1581948-001
To be published
|
|
1YD5
 
 | | Crystal structure of the GIY-YIG N-terminal endonuclease domain of UvrC from Thermotoga maritima: Point mutant N88A bound to its catalytic divalent cation | | Descriptor: | MANGANESE (II) ION, UvrABC system protein C | | Authors: | Truglio, J.J, Rhau, B, Croteau, D.L, Wang, L, Skorvaga, M, Karakas, E, DellaVecchia, M.J, Wang, H, Van Houten, B, Kisker, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the first incision reaction during nucleotide excision repair
Embo J., 24, 2005
|
|
