3IJQ
 
 | | Structure of dipeptide epimerase from Bacteroides thetaiotaomicron complexed with L-Ala-D-Glu; productive substrate binding. | | Descriptor: | ALANINE, D-GLUTAMIC ACID, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-08-04 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DFD
 
 | | Crystal structure of had family enzyme bt-2542 (target efi-501088) from bacteroides thetaiotaomicron, magnesium complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative haloacid dehalogenase-like hydrolase, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-23 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Protein Bt-2542 from Bacteroides Thetaiotaomicron (Target Efi-501088)
To be Published
|
|
4DO7
 
 | | Crystal structure of an amidohydrolase (cog3618) from burkholderia multivorans (target efi-500235) with bound zn, space group c2 | | Descriptor: | Amidohydrolase 2, SULFATE ION, ZINC ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N.F, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-09 | | Release date: | 2012-02-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an amidohydrolase (cog3618) from burkholderia multivorans (target efi-500235) with bound zn, space group c2
to be published
|
|
3IRV
 
 | | CRYSTAL STRUCTURE OF CYSTEINE HYDROLASE PSPPH_2384 FROM Pseudomonas syringae pv. phaseolicola 1448A | | Descriptor: | CYSTEINE HYDROLASE, GLYCEROL, PHOSPHATE ION | | Authors: | Patskovsky, Y, Malashkevich, V, Toro, R, Foti, R, Dickey, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-24 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF CYSTEINE HYDROLASE PSPPH_2384 FROM Pseudomonas syringae
To be Published
|
|
4DVJ
 
 | | Crystal structure of a putative zinc-dependent alcohol dehydrogenase protein from Rhizobium etli CFN 42 | | Descriptor: | Putative zinc-dependent alcohol dehydrogenase protein | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hellerich, B, Kar, A, Lafleur, J, Siedel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-23 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of a putative zinc-dependent alcohol dehydrogenase protein from Rhizobium etli CFN 42
To be Published
|
|
4DUP
 
 | | Crystal Structure of a quinone oxidoreductase from Rhizobium etli CFN 42 | | Descriptor: | quinone oxidoreductase | | Authors: | Kumaran, D, Rice, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-22 | | Release date: | 2012-03-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of a quinone oxidoreductase from Rhizobium etli CFN 42
To be Published
|
|
4DI9
 
 | | CRYSTAL STRUCTURE OF THE D248A mutant of 2-PYRONE-4,6-DICARBOXYLIC ACID HYDROLASE FROM SPHINGOMONAS PAUCIMOBILIS complexed with substrate at pH 6.5 | | Descriptor: | (1E,3Z)-4-hydroxybuta-1,3-diene-1,2,4-tricarboxylic acid, 2-pyrone-4,6-dicarbaxylate hydrolase, ACETATE ION | | Authors: | Malashkevich, V.N, Toro, R, Hobbs, M.E, Raushel, F.M, Almo, S.C. | | Deposit date: | 2012-01-11 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and Catalytic Mechanism of LigI: Insight into the Amidohydrolase Enzymes of cog3618 and Lignin Degradation.
Biochemistry, 51, 2012
|
|
3IJD
 
 | | Uncharacterized protein Cthe_2304 from Clostridium thermocellum binds two copies of 5-methyl-5,6,7,8-tetrahydrofolic acid | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, uncharacterized protein | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Miller, S, Ozyurt, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-04 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Uncharacterized protein Cthe_2304 from Clostridium thermocellum binds two copies of 5-methyl-5,6,7,8-tetrahydrofolic acid
To be Published
|
|
4DNH
 
 | | Crystal structure of hypothetical protein SMc04132 from Sinorhizobium meliloti 1021 | | Descriptor: | GLYCEROL, Uncharacterized protein | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-08 | | Release date: | 2012-03-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of hypothetical protein SMc04132 from Sinorhizobium meliloti 1021
To be Published
|
|
3IP3
 
 | | Structure of putative oxidoreductase (TM_0425) from Thermotoga maritima | | Descriptor: | Oxidoreductase, putative, SULFATE ION | | Authors: | Ramagopal, U.A, Morano, C, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-16 | | Release date: | 2009-09-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure of putative oxidoreductase (TM_0425) from Thermotoga maritima
To be Published
|
|
3ISA
 
 | | CRYSTAL STRUCTURE OF putative enoyl-CoA hydratase/isomerase FROM Bordetella parapertussis | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative enoyl-CoA hydratase/isomerase | | Authors: | Patskovsky, Y, Malashkevich, V, Toro, R, Foti, R, Dickey, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-25 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | CRYSTAL STRUCTURE OF enoyl-CoA hydratase FROM Bordetella parapertussis
To be Published
|
|
3GDK
 
 | | Crystal structure of the orotidine 5'-monophosphate decarboxylase from Saccharomyces cerevisiae | | Descriptor: | Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-02-24 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
3G5L
 
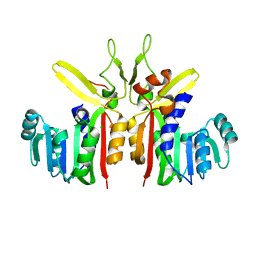 | | Crystal structure of putative S-adenosylmethionine dependent methyltransferase from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, Putative S-adenosylmethionine dependent methyltransferase | | Authors: | Patskovsky, Y, Sampathkumar, P, Gilmore, M, Miller, S, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of S-Adenosylmethionine Dependent Methyltransferase from Listeria Monocytogenes
To be Published
|
|
4DWD
 
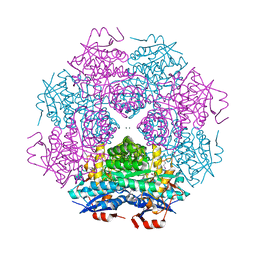 | | Crystal structure of mandelate racemase/muconate lactonizing protein from Paracoccus denitrificans PD1222 complexed with magnesium | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme, ... | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-24 | | Release date: | 2012-03-14 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of mandelate racemase/muconate lactonizing protein from Paracoccus denitrificans PD1222 complexed with magnesium
To be Published
|
|
4DN7
 
 | | CRYSTAL STRUCTURE OF putative ABC transporter, ATP-binding protein from Methanosarcina mazei Go1 | | Descriptor: | ABC transporter, ATP-binding protein, PENTAETHYLENE GLYCOL | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-08 | | Release date: | 2012-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF putative ABC transporter, ATP-binding protein from Methanosarcina mazei Go1
To be Published
|
|
3G7U
 
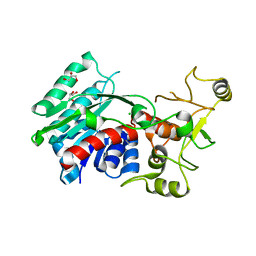 | | Crystal structure of putative DNA modification methyltransferase encoded within prophage Cp-933R (E.coli) | | Descriptor: | CHLORIDE ION, Cytosine-specific methyltransferase, GLYCEROL | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Gilmore, M, Iizuka, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-10 | | Release date: | 2009-02-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of DNA Modification Methyltransferase Encoded within Prophage Cp-933R (E.coli)
To be Published
|
|
4F3C
 
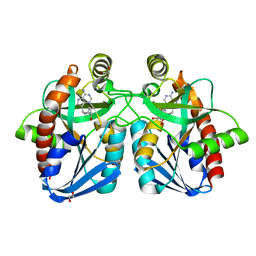 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with butyl-thio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Haapalainen, A.M, Thomas, K, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-05-09 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with butyl-thio-DADMe-Immucillin-A
Structure, 21, 2013
|
|
3GD5
 
 | | Crystal structure of ornithine carbamoyltransferase from Gloeobacter violaceus | | Descriptor: | Ornithine carbamoyltransferase | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Ramagopal, U.A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-23 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of ornithine carbamoyltransferase from Gloeobacter violaceus
To be Published
|
|
3GDT
 
 | | Crystal structure of the D91N mutant of the orotidine 5'-monophosphate decarboxylase from Saccharomyces cerevisiae complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-02-24 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
3HK9
 
 | | Crystal structure of uronate isomerase from Bacillus halodurans complexed with zinc and D-Glucuronate | | Descriptor: | CARBONATE ION, CHLORIDE ION, D-glucuronic acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Nguyen, T.T, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-05-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The mechanism of the reaction catalyzed by uronate isomerase illustrates how an isomerase may have evolved from a hydrolase within the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3HK7
 
 | | Crystal structure of uronate isomerase from Bacillus halodurans complexed with zinc and D-Arabinarate, monoclinic crystal form | | Descriptor: | CARBONATE ION, CHLORIDE ION, D-arabinaric acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Nguyen, T.T, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-05-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanism of the reaction catalyzed by uronate isomerase illustrates how an isomerase may have evolved from a hydrolase within the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3I4F
 
 | | Structure of putative 3-oxoacyl-reductase from bacillus thuringiensis | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase | | Authors: | Ramagopal, U.A, Kim, J, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of putative 3-oxoacyl-reductase from bacillus thuringiensis
To be published
|
|
3HMU
 
 | | Crystal structure of a class III aminotransferase from Silicibacter pomeroyi | | Descriptor: | Aminotransferase, class III, CHLORIDE ION, ... | | Authors: | Toro, R, Bonanno, J.B, Ramagopal, U, Freeman, J, Bain, K.T, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-29 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a class III aminotransferase from Silicibacter pomeroyi
To be Published
|
|
3IAN
 
 | | Crystal structure of a chitinase from Lactococcus lactis subsp. lactis | | Descriptor: | 1,2-ETHANEDIOL, Chitinase, SODIUM ION | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Ozyurt, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-14 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a chitinase from Lactococcus lactis subsp. lactis
To be Published
|
|
3IBM
 
 | | CRYSTAL STRUCTURE OF cupin 2 domain-containing protein Hhal_0468 FROM Halorhodospira halophila | | Descriptor: | Cupin 2, conserved barrel domain protein, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-16 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF cupin 2 domain-containing PROTEIN Hhal_0468 FROM Halorhodospira halophila
To be Published
|
|
