8HC5
 
 | | SARS-CoV-2 wildtype S1 in complex with YB9-258 Fab and R1-32 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, Heavy chain of YB9-258 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HCB
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 3 YB13-292 Fabs (2 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC7
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) complex with YB9-258 Fab, focused refinement of RBD-dimer region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain variable region of YB9-258, Light chain variable region of YB9-258, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC8
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with YB13-292 Fab, focused refinement of Fab region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, Light chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC3
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 2 YB9-258 Fabs (2 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB9-258, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC4
 
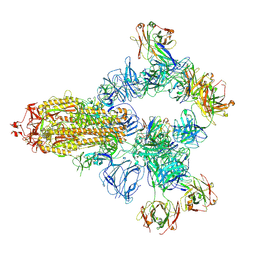 | | SARS-CoV-2 wildtype spike trimer (6P) in complex with 3 YB9-258 Fabs and 3 R1-32 Fabs (3 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HCA
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 3 YB13-292 Fabs (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC6
 
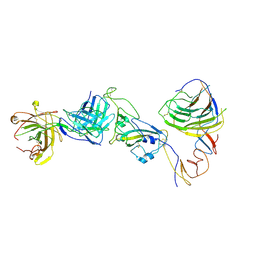 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with YB9-258 Fab, focused refinement of Fab region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB9-258 Fab, Light chain of YB9-258, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC9
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 3 YB13-292 Fabs (3 RBD down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (6.03 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC2
 
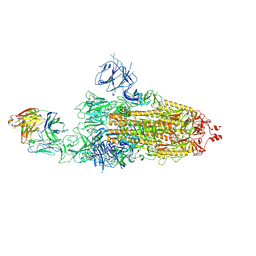 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 1 YB9-258 Fab (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB9-258 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (6.21 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
7XST
 
 | |
7XMX
 
 | | Cryo-EM structure of SARS-CoV-2 spike glycoprotein in complex with three F61 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F61 heavy chain, F61 light chain, ... | | Authors: | Wang, X, Li, X. | | Deposit date: | 2022-04-27 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis of a two-antibody cocktail exhibiting highly potent and broadly neutralizing activities against SARS-CoV-2 variants including diverse Omicron sublineages.
Cell Discov, 8, 2022
|
|
9F0H
 
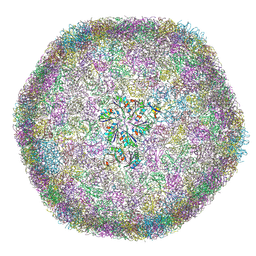 | | cryo-EM structure of carboxysomal mini-shell icosahedral assembly from co-expression of CsoS1C, CsoS4A, and CsoS2-C (T = 9) | | Descriptor: | Carboxysome assembly protein CsoS2B, Carboxysome shell protein CsoS1C, Carboxysome shell vertex protein CsoS4A, ... | | Authors: | Wang, P, Marles-Wright, J, Liu, L.N. | | Deposit date: | 2024-04-16 | | Release date: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (1.8 Å) | | Cite: | Molecular principles of the assembly and construction of a carboxysome shell.
Sci Adv, 10, 2024
|
|
9JQG
 
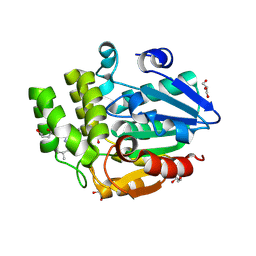 | | Crystal structure of rice DWARF14 in complex with Cyclo(L-Leu-L-Pro) | | Descriptor: | (3~{S},8~{a}~{S})-3-(2-methylpropyl)-2,3,6,7,8,8~{a}-hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, 1,3-PROPANDIOL, 1,4-BUTANEDIOL, ... | | Authors: | Wang, Q.X, Feng, Q.X, Wang, B, Bai, Y. | | Deposit date: | 2024-09-27 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Root microbiota regulates tiller number in rice.
Cell, 188, 2025
|
|
3THB
 
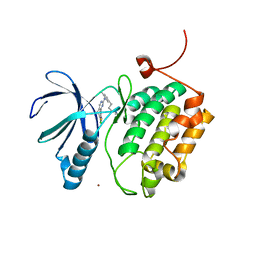 | | Structure of PLK1 kinase domain in complex with a benzolactam-derived inhibitor | | Descriptor: | 9-chloro-2-({5-[3-(dimethylamino)propyl]-2-methylpyridin-3-yl}amino)-5,7-dihydro-6H-pyrimido[5,4-d][1]benzazepine-6-thi one, Serine/threonine-protein kinase PLK1, ZINC ION | | Authors: | Sintchak, M.D. | | Deposit date: | 2011-08-18 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of a Potent and Orally Bioavailable Benzolactam-Derived Inhibitor of Polo-Like Kinase 1 (MLN0905).
J.Med.Chem., 55, 2012
|
|
5D1L
 
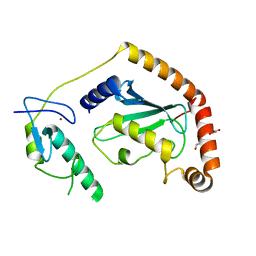 | | Crystal Structure of UbcH5B in Complex with the RING-U5BR Fragment of AO7 (Y165A) | | Descriptor: | DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase RNF25, OXALATE ION, ... | | Authors: | Liang, Y.-H, Li, S, Weissman, A.M, Ji, X. | | Deposit date: | 2015-08-04 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.618 Å) | | Cite: | Insights into Ubiquitination from the Unique Clamp-like Binding of the RING E3 AO7 to the E2 UbcH5B.
J.Biol.Chem., 290, 2015
|
|
4GUS
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P3221 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUR
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P21 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUT
 
 | | Crystal structure of LSD2-NPAC | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
6LRB
 
 | |
6IGD
 
 | | Crystal structure of HPV58/33 chimeric L1 pentamer | | Descriptor: | Major capsid protein L1 | | Authors: | Li, Z.H, Song, S, He, M.Z, Gu, Y, Li, S.W. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational design of a triple-type human papillomavirus vaccine by compromising viral-type specificity.
Nat Commun, 9, 2018
|
|
6IGF
 
 | | Crystal structure of Human Papillomavirus type 52 pentamer | | Descriptor: | Major capsid protein L1 | | Authors: | Li, Z.H, Song, S, He, M.Z, Gu, Y, Li, S.W. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Rational design of a triple-type human papillomavirus vaccine by compromising viral-type specificity.
Nat Commun, 9, 2018
|
|
6IGE
 
 | | Crystal structure of Human Papillomavirus type 33 pentamer | | Descriptor: | Major capsid protein L1 | | Authors: | Li, Z.H, Song, S, He, M.Z, Gu, Y, Li, S.W. | | Deposit date: | 2018-09-25 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Rational design of a triple-type human papillomavirus vaccine by compromising viral-type specificity.
Nat Commun, 9, 2018
|
|
4PXQ
 
 | | Crystal structure of D-glucuronyl C5-epimerase in complex with heparin hexasaccharide | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, D-glucuronyl C5 epimerase B | | Authors: | Ke, J, Qin, Y, Gu, X, Tan, J, Brunzelle, J.S, Xu, H.E, Ding, K. | | Deposit date: | 2014-03-24 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Study of d-Glucuronyl C5-epimerase.
J.Biol.Chem., 290, 2015
|
|
8J9A
 
 | |
