7XUV
 
 | | Crystal structure of RPA70N-RMI1 fusion | | Descriptor: | RecQ-mediated genome instability protein 1, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Wu, Y.Y, Zang, N, Fu, W.M, Zhou, C. | | Deposit date: | 2022-05-20 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of human RPA70N association with DNA damage response proteins.
Elife, 12, 2023
|
|
7XUW
 
 | | Crystal structure of RPA70N-BLMp2 fusion | | Descriptor: | fusion protein of Replication protein A 70 kDa DNA-binding subunit and Bloom syndrome protein | | Authors: | Wu, Y.Y, Zang, N, Fu, W.M, Zhou, C. | | Deposit date: | 2022-05-20 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of human RPA70N association with DNA damage response proteins.
Elife, 12, 2023
|
|
7DME
 
 | | Solution structure of human Aha1 | | Descriptor: | Activator of 90 kDa heat shock protein ATPase homolog 1 | | Authors: | Hu, H, Zhou, C, Zhang, N. | | Deposit date: | 2020-12-03 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Aha1 Exhibits Distinctive Dynamics Behavior and Chaperone-Like Activity.
Molecules, 26, 2021
|
|
7DMD
 
 | |
8JP3
 
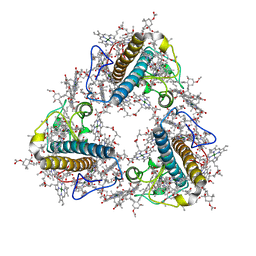 | | FCP trimer in diatom Thalassiosira pseudonana | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, CHLOROPHYLL A, ... | | Authors: | Feng, Y, Li, Z, Zhou, C.C, Liu, C, Shen, J.R, Wang, W. | | Deposit date: | 2023-06-10 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural and spectroscopic insights into fucoxanthin chlorophyll a/c-binding proteins of diatoms in diverse oligomeric states.
Plant Commun., 2024
|
|
2LMG
 
 | |
2LGW
 
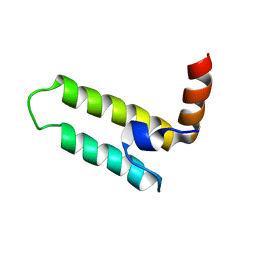 | | Solution Structure of the J Domain of HSJ1a | | Descriptor: | DnaJ homolog subfamily B member 2 | | Authors: | Zhou, C, Gao, X, Cao, C, Hu, H. | | Deposit date: | 2011-08-02 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C-terminal helices of heat shock protein 70 are essential for J-domain binding and ATPase activation.
J.Biol.Chem., 287, 2012
|
|
5XOI
 
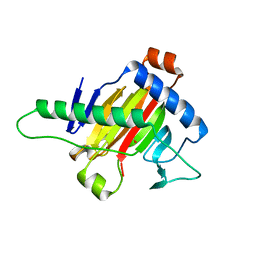 | | The structure of OsALKBH1 | | Descriptor: | MANGANESE (II) ION, Oxidoreductase, 2OG-Fe oxygenase family protein, ... | | Authors: | Wang, C, Guo, Y, Zeng, Z. | | Deposit date: | 2017-05-28 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and analysis of adenine N6-methylation sites in the rice genome.
Nat Plants, 4, 2018
|
|
5XEG
 
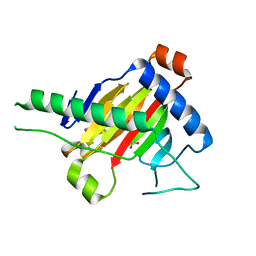 | | The structure of OsALKBH1 | | Descriptor: | 2-OXOGLUTARIC ACID, MANGANESE (II) ION, Oxidoreductase, ... | | Authors: | Wang, C, Guo, Y, Zeng, Z. | | Deposit date: | 2017-04-05 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and analysis of adenine N6-methylation sites in the rice genome.
Nat Plants, 4, 2018
|
|
2JNH
 
 | | Solution Structure of the UBA Domain from Cbl-b | | Descriptor: | E3 ubiquitin-protein ligase CBL-B | | Authors: | Zhou, C, Zhou, Z, Lin, D, Hu, H. | | Deposit date: | 2007-01-24 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Differential ubiquitin binding of the UBA domains from human c-Cbl and Cbl-b: NMR structural and biochemical insights
Protein Sci., 17, 2008
|
|
8H8Y
 
 | | Crystal structure of AbHheG from Acidimicrobiia bacterium | | Descriptor: | GLYCEROL, alpha/beta hydrolase | | Authors: | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | Deposit date: | 2022-10-24 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
8HQP
 
 | | Crystal structure of AbHheG mutant from Acidimicrobiia bacterium | | Descriptor: | AbHheG_m | | Authors: | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | Deposit date: | 2022-12-13 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
6JZE
 
 | | Crystal structure of VASH2-SVBP complex with the magic triangle I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 2 | | Authors: | Chen, Z, Ling, Y, Zeyuan, G, Zhu, L. | | Deposit date: | 2019-05-01 | | Release date: | 2019-08-07 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis of tubulin detyrosination by VASH2/SVBP heterodimer.
Nat Commun, 10, 2019
|
|
6JZD
 
 | | Crystal structure of peptide-bound VASH2-SVBP complex | | Descriptor: | GLU-GLY-GLU-GLU-TYR, Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 2 | | Authors: | Chen, Z, Ling, Y, Zeyuan, G, Zhu, L. | | Deposit date: | 2019-05-01 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.479 Å) | | Cite: | Structural basis of tubulin detyrosination by VASH2/SVBP heterodimer.
Nat Commun, 10, 2019
|
|
6JZC
 
 | | Structural basis of tubulin detyrosination | | Descriptor: | GLYCEROL, Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 2 | | Authors: | Chen, Z, Ling, Y, Zeyuan, G, Zhu, L. | | Deposit date: | 2019-05-01 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural basis of tubulin detyrosination by VASH2/SVBP heterodimer.
Nat Commun, 10, 2019
|
|
6KCZ
 
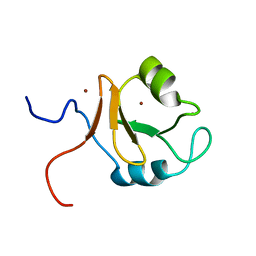 | |
2LJ1
 
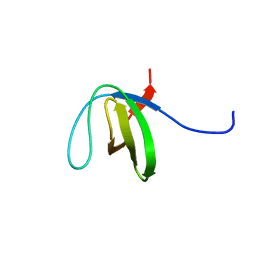 | | The third SH3 domain of R85FL with ataxin-7 PRR | | Descriptor: | Sorbin and SH3 domain-containing protein 1 | | Authors: | Jiang, Y, Hu, H. | | Deposit date: | 2011-09-02 | | Release date: | 2012-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure Basis for the Recognition of Ataxin-7 PRR with R85FL SH3 Domain
To be Published
|
|
2LJ0
 
 | | The third SH3 domain of R85FL | | Descriptor: | Sorbin and SH3 domain-containing protein 1 | | Authors: | Jiang, Y, Hu, H. | | Deposit date: | 2011-09-02 | | Release date: | 2012-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure Basis for the Recognition of Ataxin-7 PRR with R85FL SH3 Domain
To be Published
|
|
6M1J
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12x | | Descriptor: | 1-[5-[6-fluoranyl-8-(methylamino)-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]cyclopropane-1-carboxylic acid, DIMETHYL SULFOXIDE, DNA gyrase subunit B, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2020-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
6M1S
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12o | | Descriptor: | 3-[5-[8-(ethylamino)-6-fluoranyl-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]oxy-2,2-dimethyl-propanoic acid, CHLORIDE ION, DNA gyrase subunit B, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2020-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
7CMW
 
 | | Complex structure of PARP1 catalytic domain with pamiparib | | Descriptor: | (2R)-14-fluoro-2-methyl-6,9,10,19-tetrazapentacyclo[14.2.1.02,6.08,18.012,17]nonadeca-1(18),8,12(17),13,15-pentaen-11-one, GLYCEROL, Poly [ADP-ribose] polymerase 1 | | Authors: | Feng, Y.C, Peng, H, Hong, Y, Liu, Y. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Pamiparib (BGB-290), a Potent and Selective Poly (ADP-ribose) Polymerase (PARP) Inhibitor in Clinical Development.
J.Med.Chem., 63, 2020
|
|
3EVI
 
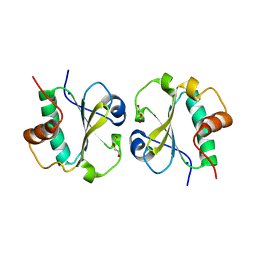 | | Crystal structure of the thioredoxin-fold domain of human phosducin-like protein 2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Phosducin-like protein 2, SODIUM ION | | Authors: | Lou, X, Bao, R, Zhou, C, Chen, Y. | | Deposit date: | 2008-10-13 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the thioredoxin-fold domain of human phosducin-like protein 2
Acta Crystallogr.,Sect.F, 65, 2009
|
|
5MWG
 
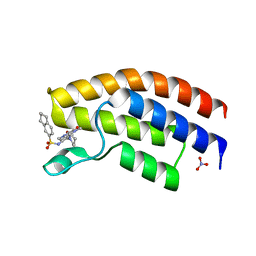 | | Crystal structure of the human BRPF1 bromodomain in complex with BZ091 | | Descriptor: | NITRATE ION, Peregrin, ~{N}-[1,4-dimethyl-2,3-bis(oxidanylidene)-7-piperidin-1-yl-quinoxalin-6-yl]-5,6,7,8-tetrahydronaphthalene-2-sulfonamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-01-18 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based discovery of selective BRPF1 bromodomain inhibitors.
Eur J Med Chem, 155, 2018
|
|
5MWH
 
 | | Crystal structure of the human BRPF1 bromodomain in complex with BZ089 | | Descriptor: | NITRATE ION, Peregrin, ~{N}-[1,4-dimethyl-7-morpholin-4-yl-2,3-bis(oxidanylidene)quinoxalin-6-yl]-4-(2-methylpropyl)benzenesulfonamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-01-18 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-based discovery of selective BRPF1 bromodomain inhibitors.
Eur J Med Chem, 155, 2018
|
|
5MWZ
 
 | |
