5DFJ
 
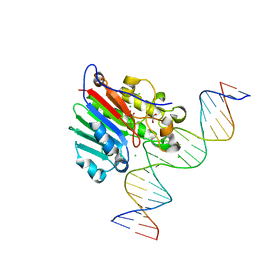 | | Human APE1 E96Q/D210N mismatch substrate complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*(3DR)P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-08-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Capturing snapshots of APE1 processing DNA damage.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5DFI
 
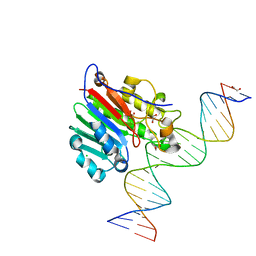 | | Human APE1 phosphorothioate substrate complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(OMC)P*(48Z)P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-08-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Capturing snapshots of APE1 processing DNA damage.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5DG0
 
 | |
5DFF
 
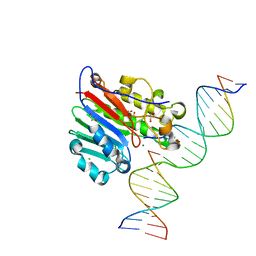 | | Human APE1 product complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-08-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Capturing snapshots of APE1 processing DNA damage.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5BOL
 
 | | DNA polymerase beta ternary complex with a templating 5ClC and incoming dGTP analog | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]guanosine, DNA (5'-D(*CP*CP*GP*AP*CP*(CDO)P*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-05-27 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Intrinsic mutagenic properties of 5-chlorocytosine: A mechanistic connection between chronic inflammation and cancer.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5BPC
 
 | | DNA polymerase beta ternary complex with a templating 5ClC and incoming dATP analog | | Descriptor: | 2'-deoxy-5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]adenosine, DNA (5'-D(*CP*CP*GP*AP*CP*(CDO)P*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-05-28 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intrinsic mutagenic properties of 5-chlorocytosine: A mechanistic connection between chronic inflammation and cancer.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5BOM
 
 | | DNA polymerase beta binary complex with a templating 5ClC | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*(4U3)P*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-05-27 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0002 Å) | | Cite: | Intrinsic mutagenic properties of 5-chlorocytosine: A mechanistic connection between chronic inflammation and cancer.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6BEM
 
 | | Ternary complex crystal structure of DNA polymerase Beta with S-isomer of beta-gamma-CHCL-dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 5'-O-[(R)-{[(R)-[(S)-chloro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-2'-deoxycytidine, CHLORIDE ION, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2017-10-25 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
2BPC
 
 | | CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA: EVIDENCE FOR A COMMON POLYMERASE MECHANISM | | Descriptor: | DNA POLYMERASE BETA, MANGANESE (II) ION | | Authors: | Sawaya, M.R, Pelletier, H, Kumar, A, Wilson, S.H, Kraut, J. | | Deposit date: | 1994-07-08 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism.
Science, 264, 1994
|
|
2BPF
 
 | | STRUCTURES OF TERNARY COMPLEXES OF RAT DNA POLYMERASE BETA, A DNA TEMPLATE-PRIMER, AND DDCTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*GP*GP*CP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*CP*GP*CP*CP*G)-3'), ... | | Authors: | Pelletier, H, Sawaya, M.R, Kumar, A, Wilson, S.H, Kraut, J. | | Deposit date: | 1994-05-19 | | Release date: | 1994-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP.
Science, 264, 1994
|
|
2BPG
 
 | | STRUCTURES OF TERNARY COMPLEXES OF RAT DNA POLYMERASE BETA, A DNA TEMPLATE-PRIMER, AND DDCTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*GP*GP*CP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*CP*GP*CP*CP*G)-3'), ... | | Authors: | Pelletier, H, Sawaya, M.R, Kumar, A, Wilson, S.H, Kraut, J. | | Deposit date: | 1994-05-19 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP.
Science, 264, 1994
|
|
6BEL
 
 | | Ternary complex crystal structure of DNA polymerase Beta with R-isomer of beta-gamma-CHF-dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-deoxy-5'-O-[(R)-{[(R)-[(R)-fluoro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]cytidine, CHLORIDE ION, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2017-10-25 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CRB
 
 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CF2, beta, gamma dATP analogue | | Descriptor: | 9-{2-deoxy-5-O-[(S)-{[(S)-[difluoro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-alpha-D-erythro-pentofuranosyl}-9H-purin-6-amine, DNA polymerase beta, Downstream Primer Strand, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CR7
 
 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CHF, beta, gamma dATP analogue | | Descriptor: | 9-{2-deoxy-5-O-[(R)-{[(R)-[(R)-fluoro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-alpha-D-erythro-pentofuranosyl}-9H-purin-6-amine, CHLORIDE ION, DNA polymerase beta, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-16 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CR4
 
 | |
6CTL
 
 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CHCL-R/S isomers, beta, gamma dTTP analogue | | Descriptor: | 5'-O-[(R)-{[(R)-[(R)-chloro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]thymidine, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CR5
 
 | |
6CTW
 
 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CCL2, beta, gamma dCTP analogue | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 4-amino-1-{2-deoxy-5-O-[(R)-{[(R)-[dichloro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-alpha-L-threo-pentofuranosyl}pyrimidin-2(1H)-one, CHLORIDE ION, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CTQ
 
 | |
6CTM
 
 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CHCL(R-isomer), beta, gamma dTTP analogue | | Descriptor: | 5'-O-[(R)-{[(R)-[(R)-chloro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]thymidine, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CR8
 
 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CHCL (R & S isomers), beta, gamma dATP analogue | | Descriptor: | 5'-O-[(R)-{[(R)-[(R)-chloro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-2'-deoxyadenosine, CHLORIDE ION, DNA polymerase beta, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-16 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CR6
 
 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CH-CH3, beta, gamma dATP analogue | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy({(R)-hydroxy[(1S)-1-phosphonoethyl]phosphoryl}oxy)phosphoryl]adenosine, DNA polymerase beta, Downstream Primer Strand, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-16 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CTN
 
 | |
6CRC
 
 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CCL2, beta, gamma dATP analogue | | Descriptor: | 2'-deoxy-5'-O-[(R)-{[(R)-[dichloro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]adenosine, DNA polymerase beta, Downstream Primer Strand, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-16 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CTP
 
 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CH2, beta, gamma dTTP analogue | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DOC))-3'), ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
