2KKZ
 
 | | Solution NMR structure of the monomeric W187R mutant of A/Udorn NS1 effector domain. Northeast Structural Genomics target OR8C[W187R]. | | Descriptor: | Non-structural protein NS1 | | Authors: | Aramini, J.M, Ma, L, Lee, H, Zhao, L, Cunningham, K, Ciccosanti, C, Janjua, H, Fang, Y, Xiao, R, Krug, R.M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-29 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the monomeric W187R mutant of A/Udorn NS1 effector domain. Northeast Structural Genomics target OR8C[W187R].
To be Published
|
|
2NWB
 
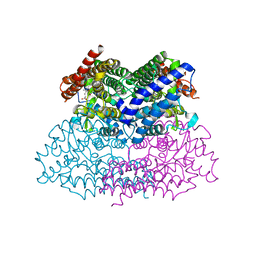 | | Crystal Structure of a Putative 2,3-dioxygenase (SO4414) from Shewanella oneidensis in complex with ferric heme. Northeast Structural Genomics Target SoR52. | | Descriptor: | Conserved domain protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Forouhar, F, Anderson, J.L.R, Mowat, C.G, Hussain, A, Seetharaman, J, Bruckmann, C, Thackray, S.J, Khan, N, Cunningham, K, Janjua, H, Zhao, L, Xiao, R, Ma, L.C, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Champman, S.K, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2NYS
 
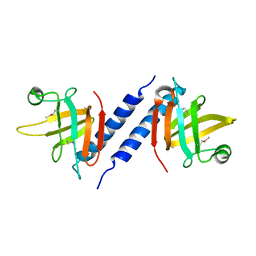 | | X-ray Crystal Structure of Protein AGR_C_3712 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR88. | | Descriptor: | AGR_C_3712p | | Authors: | Vorobiev, S.M, Abashidze, M, Seetharaman, J, Zhao, L, Ma, L.C, Cunningham, K, Nwosu, C, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-21 | | Release date: | 2006-12-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the hypothetical protein AGR_C_3712 from Agrobacterium tumefaciens.
To be Published
|
|
2O8S
 
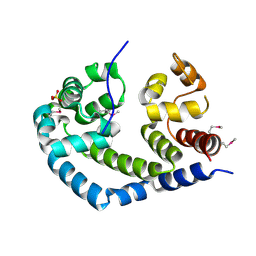 | | X-ray Crystal Structure of Protein AGR_C_984 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR120. | | Descriptor: | AGR_C_984p, SULFATE ION | | Authors: | Seetharaman, J, Forouhar, F, Su, M, Zhao, L, Cunningham, K, Ma, L.-C, Janjua, H, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-12 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Hypothetical Protein AGR_C_984 from Agrobacterium tumefaciens, Northeast Structural Genomics Target AtR120.
To be Published
|
|
2NW8
 
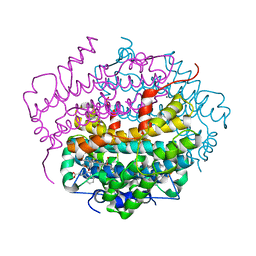 | | Crystal Structure of Tryptophan 2,3-dioxygenase (TDO) from Xanthomonas campestris in complex with ferrous heme and tryptophan. Northeast Structural Genomics Target XcR13. | | Descriptor: | MANGANESE (II) ION, PROTOPORPHYRIN IX CONTAINING FE, TRYPTOPHAN, ... | | Authors: | Forouhar, F, Anderson, J.L.R, Mowat, C.G, Bruckmann, C, Thackray, S.J, Seetharaman, J, Ho, C.K, Ma, L.C, Cunningham, K, Janjua, H, Zhao, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Champman, S.K, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2KFP
 
 | | Solution NMR structure of PSPTO_3016 from Pseudomonas syringae. Northeast Structural Genomics Consortium target PsR293. | | Descriptor: | PSPTO_3016 protein | | Authors: | Feldmann, E.A, Ramelot, T.A, Zhao, L, Hamilton, K, Ciccosanti, C, Xiao, R, Nair, R, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR and X-ray crystal structures of Pseudomonas syringae Pspto_3016 from protein domain family PF04237 (DUF419) adopt a "double wing" DNA binding motif.
J.Struct.Funct.Genom., 13, 2012
|
|
2P7N
 
 | | Crystal structure of the Pathogenicity island 1 effector protein from Chromobacterium violaceum. Northeast Structural Genomics Consortium (NESGC) target CvR69. | | Descriptor: | Pathogenicity island 1 effector protein | | Authors: | Benach, J, Abashidze, M, Seetharaman, J, Zhao, L, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-03-20 | | Release date: | 2007-04-03 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Pathogenicity island 1 effector protein from Chromobacterium violaceum. Northeast Structural Genomics Consortium (NESGC) target CvR69.
To be Published
|
|
2NW9
 
 | | Crystal Structure of Tryptophan 2,3-dioxygenase (TDO) from Xanthomonas campestris in complex with ferrous heme and 6-fluoro-tryptophan. Northeast Structural Genomics Target XcR13 | | Descriptor: | 6-FLUORO-L-TRYPTOPHAN, MANGANESE (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Forouhar, F, Anderson, J.L.R, Mowat, C.G, Bruckmann, C, Thackray, S.J, Seetharaman, J, Ho, C.K, Ma, L.C, Cunningham, K, Janjua, H, Zhao, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Champman, S.K, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2NW7
 
 | | Crystal Structure of Tryptophan 2,3-dioxygenase (TDO) from Xanthomonas campestris in complex with ferric heme. Northeast Structural Genomics Target XcR13 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase | | Authors: | Forouhar, F, Anderson, J.L.R, Mowat, C.G, Hussain, A, Bruckmann, C, Thackray, S.J, Seetharaman, J, Tucker, T, Ho, C.K, Ma, L.C, Cunningham, K, Janjua, H, Zhao, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Chapman, S.K, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
8DPX
 
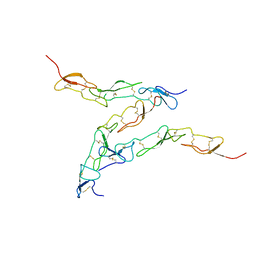 | | Preligand association structure of DR5 | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Du, G, Zhao, L, Chou, J.J. | | Deposit date: | 2022-07-17 | | Release date: | 2023-02-15 | | Method: | SOLUTION NMR | | Cite: | Autoinhibitory structure of preligand association state implicates a new strategy to attain effective DR5 receptor activation.
Cell Res., 33, 2023
|
|
2K5D
 
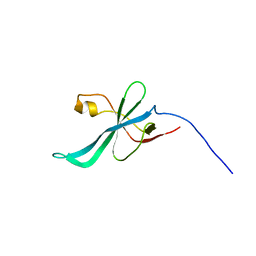 | | SOLUTION NMR STRUCTURE OF SAG0934 from Streptococcus agalactiae. NORTHEAST STRUCTURAL GENOMICS TARGET SaR32[1-108]. | | Descriptor: | uncharacterized protein SAG0934 | | Authors: | Aramini, J.M, Rossi, P, Zhao, L, Foote, E.L, Jiang, M, Xiao, R, Sharma, S, Swapna, G.VT, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-26 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | SOLUTION NMR STRUCTURE OF SAG0934 from Streptococcus agalactiae. NORTHEAST STRUCTURAL GENOMICS TARGET SaR32[1-108].
To be Published
|
|
2K5F
 
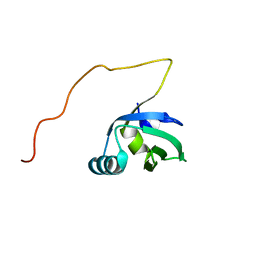 | | Solution NMR structure of FeoA protein from Chlorobium tepidum. Northeast Structural Genomics Consortium target CtR121 | | Descriptor: | Ferrous iron transport protein A | | Authors: | Eletsky, A, Sathyamoorthy, B, Mills, J.L, Zeri, A, Zhao, L, Hamilton, K, Foote, E.L, Xiao, R, Nair, R, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-27 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of FeoA protein from Chlorobium tepidum. Northeast Structural Genomics Consortium target CtR121
To be Published
|
|
2K1G
 
 | | Solution NMR structure of lipoprotein spr from Escherichia coli K12. Northeast Structural Genomics target ER541-37-162 | | Descriptor: | Lipoprotein spr | | Authors: | Aramini, J.M, Rossi, P, Zhao, L, Jiang, M, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Huang, Y.J, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-03 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the NlpC/P60 domain of lipoprotein Spr from Escherichia coli: structural evidence for a novel cysteine peptidase catalytic triad.
Biochemistry, 47, 2008
|
|
2K1S
 
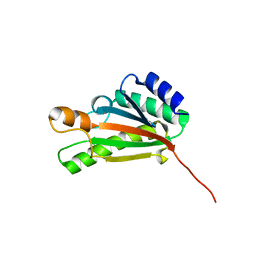 | | Solution NMR structure of the folded C-terminal fragment of YiaD from Escherichia coli. Northeast Structural Genomics Consortium target ER553. | | Descriptor: | Inner membrane lipoprotein yiaD | | Authors: | Ramelot, T.A, Zhao, L, Hamilton, K, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-14 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the folded C-terminal fragment of YiaD from Escherichia coli. Northeast Structural Genomics Consortium target ER553.
To be Published
|
|
2K3D
 
 | | Solution NMR structure of the folded 79 residue fragment of Lin0334 from Listeria innocua. Northeast Structural Genomics Consortium target LkR15 | | Descriptor: | Lin0334 protein | | Authors: | Ramelot, T.A, Zhao, L, Jiang, M, Foote, E.L, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-05-02 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the folded 79 residue fragment of Lin0334 from Listeria innocua. Northeast Structural Genomics Consortium target LkR15.
To be Published
|
|
2JVD
 
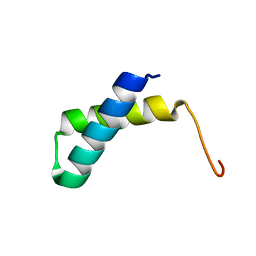 | | Solution NMR structure of the folded N-terminal fragment of UPF0291 protein ynzC from Bacillus subtilis. Northeast Structural Genomics target SR384-1-46 | | Descriptor: | UPF0291 protein ynzC | | Authors: | Aramini, J.M, Sharma, S, Huang, Y.J, Zhao, L, Owens, L.A, Stokes, K, Jiang, M, Xiao, R, Baran, M.C, Swapna, G.V.T, Acton, T.B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-18 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the SOS response protein YnzC from Bacillus subtilis.
Proteins, 72, 2008
|
|
2KCQ
 
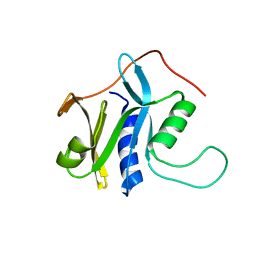 | | Solution structure of protein SRU_2040 from Salinibacter ruber (strain DSM 13855) . Northeast Structural Genomics Consortium target SrR106 | | Descriptor: | Mov34/MPN/PAD-1 family | | Authors: | Wu, Y, Eletsky, A, Zhao, L, Hua, J, Sukumaran, D, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-28 | | Release date: | 2009-02-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein SRU_2040 from Salinibacter ruber (strain DSM 13855). Northeast Structural Genomics Consortium target SrR106
To be Published
|
|
2K9Q
 
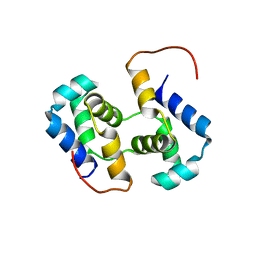 | | Solution NMR structure of HTH_XRE family transcriptional regulator BT_p548217 from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR244. | | Descriptor: | uncharacterized protein | | Authors: | Ramelot, T.A, Cort, J.R, Zhao, L, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-23 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of HTH_XRE family transcriptional regulator BT_p548217 from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR244.
To be Published
|
|
2K4Y
 
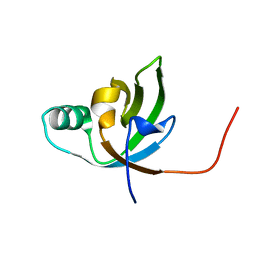 | | NMR Structure of FeoA-like protein from Clostridium acetobutylicum: Northeast Structural Genomics Consortium Target CaR178 | | Descriptor: | FeoA-like protein | | Authors: | Singarapu, K, Wu, Y, Hua, J, Sukumaran, D, Zhao, L, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Baran, M.C, Swapna, G.V.T, Acton, T, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of FeoA-like protein from Clostridium acetobutylicum: Northeast Structural Genomics Consortium Target CaR178
To be Published
|
|
2RHK
 
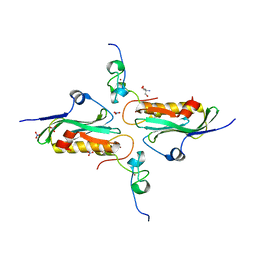 | | Crystal structure of influenza A NS1A protein in complex with F2F3 fragment of human cellular factor CPSF30, Northeast Structural Genomics Targets OR8C and HR6309A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cleavage and polyadenylation specificity factor subunit 4, NITRATE ION, ... | | Authors: | Das, K, Ma, L.-C, Xiao, R, Radvansky, B, Aramini, J, Zhao, L, Arnold, E, Krug, R.M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-10-09 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for suppression of a host antiviral response by influenza A virus.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4KWE
 
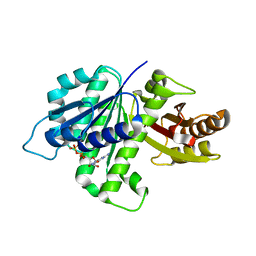 | | GDP-bound, double-stranded, curved FtsZ protofilament structure | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Li, Y, Hsin, J, Zhao, L, Cheng, Y, Shang, W, Huang, K.C, Wang, H.W, Ye, S. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | FtsZ protofilaments use a hinge-opening mechanism for constrictive force generation
Science, 341, 2013
|
|
4JBJ
 
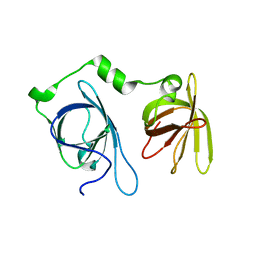 | | Structural mimicry for functional antagonism | | Descriptor: | Interferon-activable protein 202 | | Authors: | Ru, H, Ni, X, Ma, F, Zhao, L, Ding, W, Hung, L.-W, Shaw, N, Cheng, G, Liu, Z.-J. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Structural basis for termination of AIM2-mediated signaling by p202
Cell Res., 23, 2013
|
|
4JBM
 
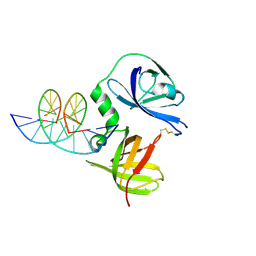 | | Structure of murine DNA binding protein bound with ds DNA | | Descriptor: | DNA (5'-D(*GP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), Interferon-inducible protein AIM2 | | Authors: | Ru, H, Ni, X, Crowley, C, Zhao, L, Ding, W, Hung, L.-W, Shaw, N, Cheng, G, Liu, Z.-J. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.218 Å) | | Cite: | Structural basis for termination of AIM2-mediated signaling by p202
Cell Res., 23, 2013
|
|
4JBK
 
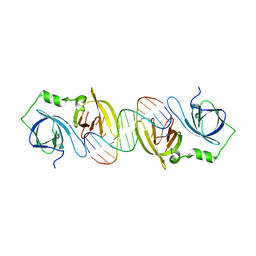 | | Molecular basis for abrogation of activation of pro-inflammatory cytokines | | Descriptor: | DNA (5'-D(P*GP*GP*AP*AP*TP*TP*AP*TP*AP*AP*TP*TP*CP*C)-3'), Interferon-activable protein 202 | | Authors: | Ru, H, Ni, X, Crowley, C, Zhao, L, Ding, W, Hung, L.-W, Shaw, N, Cheng, G, Liu, Z.-J. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.963 Å) | | Cite: | Structural basis for termination of AIM2-mediated signaling by p202
Cell Res., 23, 2013
|
|
8FEH
 
 | |
