7N2Y
 
 | |
2JGO
 
 | | Structure of the arsenated de novo designed peptide Coil Ser L9C | | Descriptor: | ARSENIC, COIL SER L9C, ZINC ION | | Authors: | Touw, D.S, Nordman, C.E, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2007-02-13 | | Release date: | 2007-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Identifying Important Structural Characteristics of Arsenic Resistance Proteins by Using Designed Three-Stranded Coiled Coils.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2JK7
 
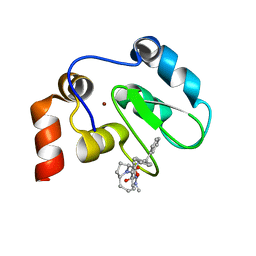 | | XIAP BIR3 bound to a Smac Mimetic | | Descriptor: | (3S,6S,7Z,10AS)-N-(DIPHENYLMETHYL)-6-{[(2S)-2-(METHYLIDENEAMINO)BUTANOYL]AMINO}-5-OXO-1,2,3,5,6,9,10,10A-OCTAHYDROPYRROLO[1,2-A]AZOCINE-3-CARBOXAMIDE, BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 4, ZINC ION | | Authors: | Saito, N.G, Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2008-08-21 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structure-Based Design, Synthesis, Evaluation, and Crystallographic Studies of Conformationally Constrained Smac Mimetics as Inhibitors of the X-Linked Inhibitor of Apoptosis Protein (Xiap).
J.Med.Chem., 51, 2008
|
|
1XXP
 
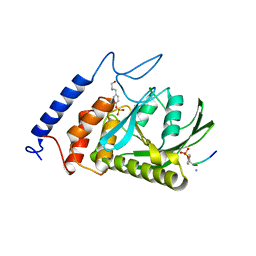 | | Yersinia YopH (residues 163-468) C403S binds phosphotyrosyl peptide at two sites | | Descriptor: | Hexapeptide ASP-ALA-ASP-GLU-PTR-CLE, Protein-tyrosine phosphatase yopH | | Authors: | Ivanov, M.I, Stuckey, J.A, Schubert, H.L, Saper, M.A, Bliska, J.B. | | Deposit date: | 2004-11-07 | | Release date: | 2005-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Two substrate-targeting sites in the Yersinia protein tyrosine phosphatase co-operate to promote bacterial virulence
Mol.Microbiol., 55, 2005
|
|
1XXV
 
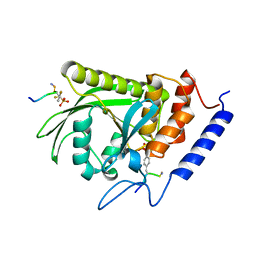 | | Yersinia YopH (residues 163-468) binds phosphonodifluoromethyl-Phe containing hexapeptide at two sites | | Descriptor: | Epidermal growth factor receptor derived peptide, Protein-tyrosine phosphatase yopH | | Authors: | Ivanov, M.I, Stuckey, J.A, Schubert, H.L, Saper, M.A, Bliska, J.B. | | Deposit date: | 2004-11-08 | | Release date: | 2005-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two substrate-targeting sites in the Yersinia protein tyrosine phosphatase co-operate to promote bacterial virulence
Mol.Microbiol., 55, 2005
|
|
4EK8
 
 | |
4EK5
 
 | |
4EK4
 
 | |
4EK6
 
 | |
4EK3
 
 | |
1LRN
 
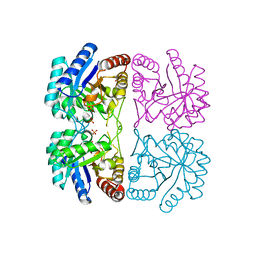 | | Aquifex aeolicus KDO8P synthase H185G mutant in complex with Cadmium | | Descriptor: | CADMIUM ION, KDO-8-phosphate synthetase, PHOSPHATE ION | | Authors: | Wang, J, Duewel, H.S, Stuckey, J.A, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2002-05-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Function of His185 in Aquifex aeolicus 3-Deoxy-D-manno-octulosonate 8-Phosphate Synthase
J.Mol.Biol., 324, 2002
|
|
1LRO
 
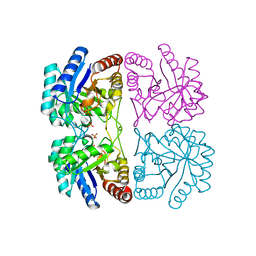 | | Aquifex aeolicus KDO8P synthase H185G mutant in complex with PEP and Cadmium | | Descriptor: | CADMIUM ION, KDO-8-phosphate synthetase, PHOSPHATE ION, ... | | Authors: | Wang, J, Duewel, H.S, Stuckey, J.A, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2002-05-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Function of His185 in Aquifex aeolicus 3-Deoxy-D-manno-octulosonate 8-Phosphate Synthase
J.Mol.Biol., 324, 2002
|
|
1LRQ
 
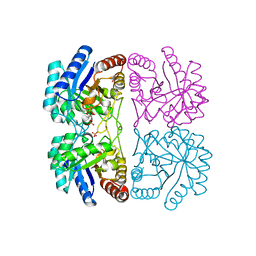 | | Aquifex aeolicus KDO8P synthase H185G mutant in complex with PEP, A5P and Cadmium | | Descriptor: | ARABINOSE-5-PHOSPHATE, CADMIUM ION, KDO-8-phosphate synthetase, ... | | Authors: | Wang, J, Duewel, H.S, Stuckey, J.A, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2002-05-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Function of His185 in Aquifex aeolicus 3-Deoxy-D-manno-octulosonate 8-Phosphate Synthase
J.Mol.Biol., 324, 2002
|
|
1LW3
 
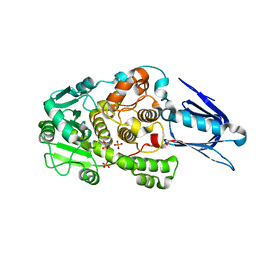 | | Crystal Structure of Myotubularin-related protein 2 complexed with phosphate | | Descriptor: | Myotubularin-related protein 2, PHOSPHATE ION | | Authors: | Begley, M.J, Taylor, G.S, Kim, S.-A, Veine, D.M, Dixon, J.E, Stuckey, J.A. | | Deposit date: | 2002-05-30 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a Phosphoinositide Phosphatase, MTMR2: Insights into Myotubular Myopathy and Charcot-Marie-Tooth Syndrome
Mol.Cell, 12, 2003
|
|
1M7R
 
 | | Crystal Structure of Myotubularin-related Protein-2 (MTMR2) Complexed with Phosphate | | Descriptor: | Myotubularin-related Protein-2, PHOSPHATE ION | | Authors: | Begley, M.J, Taylor, G.S, Kim, S.-A, Veine, D.M, Dixon, J.E, Stuckey, J.A. | | Deposit date: | 2002-07-22 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a phosphoinositide phosphatase, MTMR2: insights into myotubular myopathy and Charcot-Marie-Tooth syndrome
Mol.Cell, 12, 2003
|
|
7KMU
 
 | | Structure of WT Malaysian Banana Lectin | | Descriptor: | 1,2-ETHANEDIOL, Jacalin-type lectin domain-containing protein | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2020-11-03 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Targeted disruption of pi-pi stacking in Malaysian banana lectin reduces mitogenicity while preserving antiviral activity.
Sci Rep, 11, 2021
|
|
7KMV
 
 | | Structure of Malaysian Banana Lectin F84T | | Descriptor: | Jacalin-type lectin domain-containing protein, SULFATE ION | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2020-11-03 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeted disruption of pi-pi stacking in Malaysian banana lectin reduces mitogenicity while preserving antiviral activity.
Sci Rep, 11, 2021
|
|
6U6F
 
 | | The crystal structure of anti-apoptotic Mcl-1 protein in complex with 2, 5-substituted benzoic acid inhibitor 21 | | Descriptor: | 2-[({4-[(4-tert-butylphenyl)methyl]piperazin-1-yl}sulfonyl)amino]-5-[(2-phenylethyl)sulfanyl]benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Yang, Y, Stuckey, J.A, Nikolovska-Coleska, Z. | | Deposit date: | 2019-08-29 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery and Characterization of 2,5-Substituted Benzoic Acid Dual Inhibitors of the Anti-apoptotic Mcl-1 and Bfl-1 Proteins.
J.Med.Chem., 63, 2020
|
|
6U9K
 
 | | MLL1 SET N3861I/Q3867L bound to inhibitor 18 (TC-5153) | | Descriptor: | 5'-([(3S)-3-amino-3-carboxypropyl]{[1-(3,3-diphenylpropyl)azetidin-3-yl]methyl}amino)-5'-deoxyadenosine, GLYCEROL, Histone-lysine N-methyltransferase, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of MLL Methyltransferase.
Acs Med.Chem.Lett., 11, 2020
|
|
6U9N
 
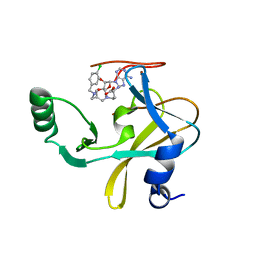 | | MLL1 SET N3861I/Q3867L bound to inhibitor 14 (TC-5139) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl]({1-[(4-chlorophenyl)methyl]azetidin-3-yl}methyl)amino}-5'-deoxyadenosine, Histone-lysine N-methyltransferase, ZINC ION | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of MLL Methyltransferase.
Acs Med.Chem.Lett., 11, 2020
|
|
6U9R
 
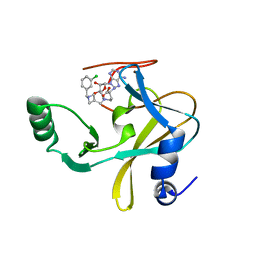 | | MLL1 SET N3861I/Q3867L bound to inhibitor 12 (TC-5140) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl]({1-[(3-chlorophenyl)methyl]azetidin-3-yl}methyl)amino}-5'-deoxyadenosine, Histone-lysine N-methyltransferase, ZINC ION | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of MLL Methyltransferase.
Acs Med.Chem.Lett., 11, 2020
|
|
5U9U
 
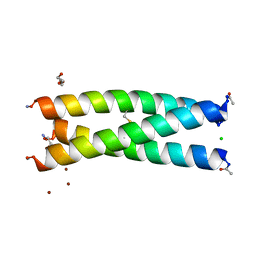 | | De Novo Three-stranded Coiled Coil Peptide Containing a Tris-thiolate Site Engineered by D-Cysteine Ligands | | Descriptor: | Apo-(CoilSer L16(DCY))3, CHLORIDE ION, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Ruckthong, L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2016-12-18 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | d-Cysteine Ligands Control Metal Geometries within De Novo Designed Three-Stranded Coiled Coils.
Chemistry, 23, 2017
|
|
5U9T
 
 | | The Tris-thiolate Zn(II)S3Cl Binding Site Engineered by D-Cysteine Ligands in de Novo Three-stranded Coiled Coil Environment | | Descriptor: | ACETATE ION, CHLORIDE ION, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Ruckthong, L, Peacock, A.F.A, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2016-12-18 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | d-Cysteine Ligands Control Metal Geometries within De Novo Designed Three-Stranded Coiled Coils.
Chemistry, 23, 2017
|
|
5UOO
 
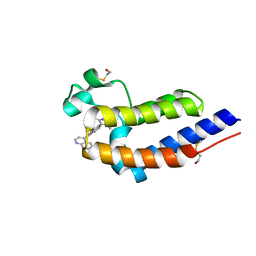 | | BRD4 bromodomain 2 in complex with CD161 | | Descriptor: | 7-(3,5-dimethyl-1,2-oxazol-4-yl)-6-methoxy-2-methyl-4-(quinolin-4-yl)-9H-pyrimido[4,5-b]indole, Bromodomain-containing protein 4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2017-02-01 | | Release date: | 2017-05-17 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-Based Discovery of 4-(6-Methoxy-2-methyl-4-(quinolin-4-yl)-9H-pyrimido[4,5-b]indol-7-yl)-3,5-dimethylisoxazole (CD161) as a Potent and Orally Bioavailable BET Bromodomain Inhibitor.
J. Med. Chem., 60, 2017
|
|
5VTB
 
 | | Crystal structure of RBBP4 bound to BCL11a peptide | | Descriptor: | B-cell lymphoma/leukemia 11A, GLYCEROL, Histone-binding protein RBBP4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2017-05-16 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the interaction between the histone methyltransferase/deacetylase subunit RBBP4/7 and the transcription factor BCL11A in epigenetic complexes.
J. Biol. Chem., 293, 2018
|
|
