2VBN
 
 | | Molecular basis of human XPC gene recognition and cleavage by engineered homing endonuclease heterodimers | | Descriptor: | 5'-D(*AP*AP*AP*AP*GP*GP*CP*AP*GP*AP)-3', 5'-D(*AP*GP*GP*AP*TP*CP*CP*TP*AP*AP)-3', 5'-D(*TP*CP*TP*GP*CP*CP*TP*TP*TP*TP *TP*TP*GP*AP)-3', ... | | Authors: | Redondo, P, Prieto, J, Munoz, I.G, Alibes, A, Stricher, F, Serrano, L, Arnould, S, Perez, C, Cabaniols, J.P, Duchateau, P, Paques, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2007-09-14 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis of Xeroderma Pigmentosum Group C DNA Recognition by Engineered Meganucleases
Nature, 456, 2008
|
|
2VBJ
 
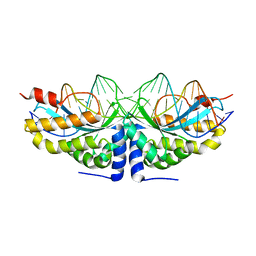 | | Molecular basis of human XPC gene recognition and cleavage by engineered homing endonuclease heterodimers | | Descriptor: | 5'-D(*TP*CP*TP*GP*CP*CP*TP*TP*TP*TP *TP*TP*GP*AP*AP*GP*GP*AP*TP*CP*CP*TP*AP*A)-3', 5'-D(*TP*TP*AP*GP*GP*AP*TP*CP*CP*TP *TP*CP*AP*AP*AP*AP*AP*AP*GP*GP*CP*AP*GP*A)-3', CALCIUM ION, ... | | Authors: | Redondo, P, Prieto, J, Munoz, I.G, Alibes, A, Stricher, F, Serrano, L, Arnould, S, Perez, C, Cabaniols, J.P, Duchateau, P, Paques, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2007-09-14 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Basis of Xeroderma Pigmentosum Group C DNA Recognition by Engineered Meganucleases
Nature, 456, 2008
|
|
2VS7
 
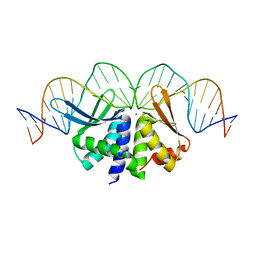 | | The crystal structure of I-DmoI in complex with DNA and Ca | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP *CP*TP*TP*AP*CP*CP*CP*GP*GP*CP*AP*AP*GP*GP*C)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP *GP*TP*AP*AP*GP*TP*TP*CP*CP*GP*GP*CP*GP*CP*G)-3', ACETATE ION, ... | | Authors: | Marcaida, M.J, Prieto, J, Redondo, P, Nadra, A.D, Alibes, A, Serrano, L, Grizot, S, Duchateau, P, Paques, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2008-04-21 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of I-Dmoi in Complex with its Target DNA Provides New Insights Into Meganuclease Engineering.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2VS8
 
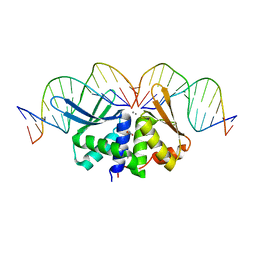 | | The crystal structure of I-DmoI in complex with DNA and Mn | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP* GP*GP*CP)-3', 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP *CP*TP*TP*AP*C)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP *GP*TP*AP*A)-3', ... | | Authors: | Marcaida, M.J, Prieto, J, Redondo, P, Nadra, A.D, Alibes, A, Serrano, L, Grizot, S, Duchateau, P, Paques, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2008-04-21 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of I-Dmoi in Complex with its Target DNA Provides New Insights Into Meganuclease Engineering.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2XE0
 
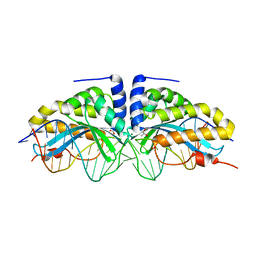 | | Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus | | Descriptor: | 24MER DNA, ACETATE ION, I-CREI V2V3 VARIANT, ... | | Authors: | Munoz, I.G, Prieto, J, Subramanian, S, Coloma, J, Redondo, P, Villate, M, Merino, N, Marenchino, M, D'Abramo, M, Gervasio, F.L, Grizot, S, Daboussi, F, Smith, J, Chion-Sotine, I, Paques, F, Duchateau, P, Alibes, A, Stricher, F, Serrano, L, Blanco, F.J, Montoya, G. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Molecular Basis of Engineered Meganuclease Targeting of the Endogenous Human Rag1 Locus
Nucleic Acids Res., 39, 2011
|
|
2VBO
 
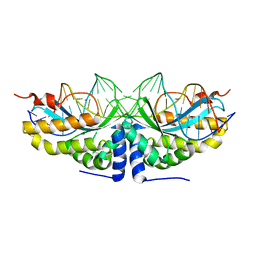 | | Molecular basis of human XPC gene recognition and cleavage by engineered homing endonuclease heterodimers | | Descriptor: | 5'-D(*TP*CP*TP*GP*CP*CP*TP*TP*TP*TP *TP*TP*GP*AP*AP*GP*GP*AP*TP*CP*CP*TP*AP*A)-3', 5'-D(*TP*TP*AP*GP*GP*AP*TP*CP*CP*TP *TP*CP*AP*AP*AP*AP*AP*AP*GP*GP*CP*AP*GP*A)-3', CALCIUM ION, ... | | Authors: | Redondo, P, Prieto, J, Munoz, I.G, Alibes, A, Stricher, F, Serrano, L, Arnould, S, Perez, C, Cabaniols, J.P, Duchateau, P, Paques, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2007-09-14 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis of Xeroderma Pigmentosum Group C DNA Recognition by Engineered Meganucleases
Nature, 456, 2008
|
|
2VNF
 
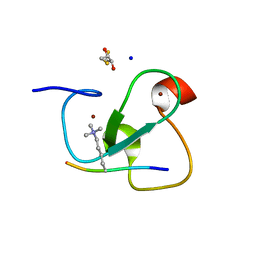 | | MOLECULAR BASIS OF HISTONE H3K4ME3 RECOGNITION BY ING4 | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, HISTONE H3, ... | | Authors: | Palacios, A, Munoz, I.G, Pantoja-Uceda, D, Marcaida, M.J, Torres, D, Martin-Garcia, J.M, Luque, I, Montoya, G, Blanco, F.J. | | Deposit date: | 2008-02-04 | | Release date: | 2008-04-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular Basis of Histone H3K4Me3 Recognition by Ing4
J.Biol.Chem., 283, 2008
|
|
5O0Y
 
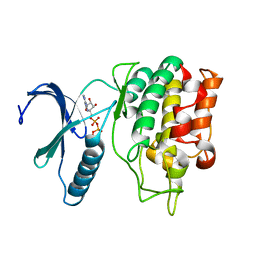 | | TLK2 kinase domain from human | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase tousled-like 2 | | Authors: | Mortuza, G.B, Montoya, G. | | Deposit date: | 2017-05-17 | | Release date: | 2018-05-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Molecular basis of Tousled-Like Kinase 2 activation.
Nat Commun, 9, 2018
|
|
6R8I
 
 | | PP4R3A EVH1 domain bound to FxxP motif | | Descriptor: | SER-LEU-PRO-PHE-THR-PHE-LYS-VAL-PRO-ALA-PRO-PRO-PRO-SER-LEU-PRO-PRO-SER, Serine/threonine-protein phosphatase 4 regulatory subunit 3A | | Authors: | Ueki, Y, Kruse, T, Weisser, M.B, Sundell, G.N, Yoo Larsen, M.S, Lopez Mendez, B, Jenkins, N.P, Garvanska, D.H, Cressey, L, Zhang, G, Davey, N, Montoya, G, Ivarsson, Y, Kettenbach, A, Nilsson, J. | | Deposit date: | 2019-04-02 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.517 Å) | | Cite: | A Consensus Binding Motif for the PP4 Protein Phosphatase.
Mol.Cell, 76, 2019
|
|
4AAE
 
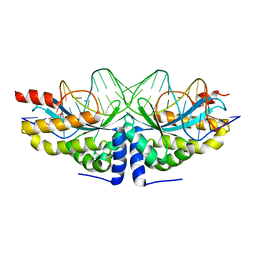 | | Crystal structure of the mutant D75N I-CreI in complex with an altered target (The four central bases, 2NN region, are composed by AGCG from 5' to 3') | | Descriptor: | 24MER DNA, DNA ENDONUCLEASE I-CREI | | Authors: | Molina, R, Redondo, P, Stella, S, Marenchino, M, D'Abramo, M, Gervasio, F.L, Epinat, J.C, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Non-Specific Protein-DNA Interactions Control I-Crei Target Binding and Cleavage.
Nucleic Acids Res., 40, 2012
|
|
4AFL
 
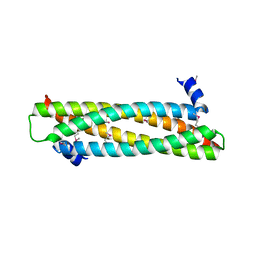 | | The crystal structure of the ING4 dimerization domain reveals the functional organization of the ING family of chromatin binding proteins. | | Descriptor: | INHIBITOR OF GROWTH PROTEIN 4 | | Authors: | Culurgioni, S, Munoz, I.G, Moreno, A, Palacios, A, Villate, M, Palmero, I, Montoya, G, Blanco, F.J. | | Deposit date: | 2012-01-19 | | Release date: | 2012-02-22 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (2.275 Å) | | Cite: | Crystal Structure of Inhibitor of Growth 4 (Ing4) Dimerization Domain Reveals Functional Organization of Ing Family of Chromatin-Binding Proteins.
J.Biol.Chem., 287, 2012
|
|
4AAF
 
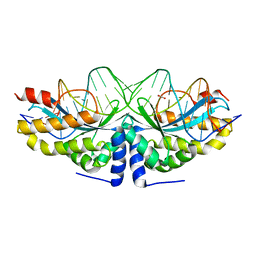 | | Crystal structure of the mutant D75N I-CreI in complex with an altered target (The four central bases, 2NN region, are composed by TGCA from 5' to 3') | | Descriptor: | 1,2-ETHANEDIOL, 24MER DNA, DNA ENDONUCLEASE I-CREI | | Authors: | Molina, R, Redondo, P, Stella, S, Marenchino, M, D'Abramo, M, Gervasio, F.L, Epinat, J.C, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-Specific Protein-DNA Interactions Control I-Crei Target Binding and Cleavage.
Nucleic Acids Res., 40, 2012
|
|
4AAD
 
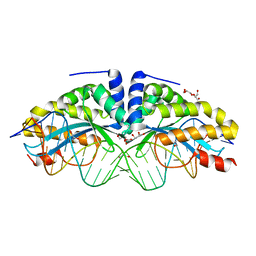 | | Crystal structure of the mutant D75N I-CreI in complex with its wild- type target in absence of metal ions at the active site (The four central bases, 2NN region, are composed by GTAC from 5' to 3') | | Descriptor: | 24MER DNA, DNA ENDONUCLEASE I-CREI, GLYCEROL | | Authors: | Molina, R, Redondo, P, Stella, S, Marenchino, M, D'Abramo, M, Gervasio, F.L, Epinat, J.C, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Non-Specific Protein-DNA Interactions Control I-Crei Target Binding and Cleavage.
Nucleic Acids Res., 40, 2012
|
|
4AQX
 
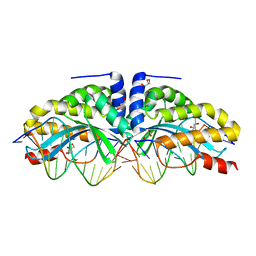 | | Crystal structure of I-CreI complexed with its target methylated at position plus 2 (in the b strand) in the presence of magnesium | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*5CM)-3', 5'-D(*GP*AP*CP*AP*GP*TP*TP*TP*GP*GP)-3', 5'-D(*GP*AP*CP*GP*TP*TP*TP*TP*GP*AP)-3', ... | | Authors: | Valton, J, Daboussi, F, Leduc, S, Redondo, P, Macmaster, R, Molina, R, Montoya, G, Duchateau, P. | | Deposit date: | 2012-04-19 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 5'-Cytosine-Phosphoguanine (Cpg) Methylation Impacts the Activity of Natural and Engineered Meganucleases.
J.Biol.Chem., 287, 2012
|
|
4AAG
 
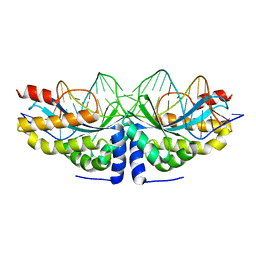 | | Crystal structure of the mutant D75N I-CreI in complex with its wild- type target in presence of Ca at the active site (The four central bases, 2NN region, are composed by GTAC from 5' to 3') | | Descriptor: | 5'-D(*TP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*AP*CP *GP*AP*CP*GP*TP*TP*TP*TP*GP*A)-3', CALCIUM ION, DNA ENDONUCLEASE I-CREI | | Authors: | Molina, R, Redondo, P, Stella, S, Marenchino, M, D'Abramo, M, Gervasio, F, Epinat, J.C, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Non-Specific Protein-DNA Interactions Control I-Crei Target Binding and Cleavage.
Nucleic Acids Res., 40, 2012
|
|
4AAB
 
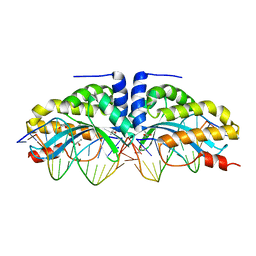 | | Crystal structure of the mutant D75N I-CreI in complex with its wild- type target (The four central bases, 2NN region, are composed by GTAC from 5' to 3') | | Descriptor: | 10MER DNA 5'-D(*GP*AP*CP*GP*TP*TP*TP*TP*GP*AP)-3', 14MER DNA 5'-D(*TP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*AP*CP)-3', DNA ENDONUCLEASE I-CREI, ... | | Authors: | Molina, R, Redondo, P, Stella, S, Marenchino, M, D'Abramo, M, Gervasio, F.L, Epinat, J.C, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-Specific Protein-DNA Interactions Control I-Crei Target Binding and Cleavage.
Nucleic Acids Res., 40, 2012
|
|
4AQU
 
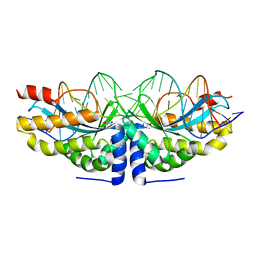 | | Crystal structure of I-CreI complexed with its target methylated at position plus 2 (in the b strand) in the presence of calcium | | Descriptor: | 5'-D(*DCP*CP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*5CMP *GP*AP*CP*GP*TP*TP*TP*TP*GP*A)-3', 5'-D(*DTP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*DAP *GP*AP*CP*AP*GP*TP*TP*TP*GP*G)-3', CALCIUM ION, ... | | Authors: | Valton, J, Daboussi, F, Leduc, S, Redondo, P, Macmaster, R, Molina, R, Montoya, G, Duchateau, P. | | Deposit date: | 2012-04-19 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 5'-Cytosine-Phosphoguanine (Cpg) Methylation Impacts the Activity of Natural and Engineered Meganucleases.
J.Biol.Chem., 287, 2012
|
|
6QZT
 
 | |
6R9R
 
 | | Crystal structure of Csx1 in complex with cyclic oligoadenylate cOA4 conformation 2 | | Descriptor: | CRISPR-associated (Cas) DxTHG family, circular RNA (5'-R(P*AP*AP*AP*A)-3') | | Authors: | Molina, R, Montoya, G, Sofos, N, Stella, S. | | Deposit date: | 2019-04-03 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Csx1-cOA4complex reveals the basis of RNA decay in Type III-B CRISPR-Cas.
Nat Commun, 10, 2019
|
|
6R7B
 
 | | Crystal structure of Csx1 in complex with cyclic oligoadenylate cOA4 conformation 1 | | Descriptor: | CRISPR-associated (Cas) DxTHG family, RNA (5'-R(P*AP*AP*AP*A)-3') | | Authors: | Molina, R, Montoya, G, Sofos, N, Stella, S. | | Deposit date: | 2019-03-28 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structure of Csx1-cOA4complex reveals the basis of RNA decay in Type III-B CRISPR-Cas.
Nat Commun, 10, 2019
|
|
6QZQ
 
 | |
8QBM
 
 | |
8QBK
 
 | |
8QBL
 
 | |
3KL4
 
 | | Recognition of a signal peptide by the signal recognition particle | | Descriptor: | Signal peptide of yeast dipeptidyl aminopeptidase B, Signal recognition 54 kDa protein | | Authors: | Janda, C.Y, Nagai, K, Li, J, Oubridge, C. | | Deposit date: | 2009-11-06 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Recognition of a signal peptide by the signal recognition particle.
Nature, 465, 2010
|
|
