3CIY
 
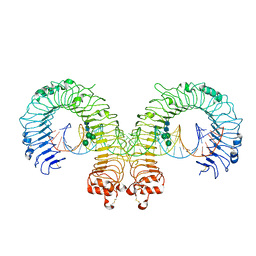 | | Mouse Toll-like receptor 3 ectodomain complexed with double-stranded RNA | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, L, Botos, I, Wang, Y, Leonard, J.N, Shiloach, J, Segal, D.M, Davies, D.R. | | Deposit date: | 2008-03-12 | | Release date: | 2008-05-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Structural basis of toll-like receptor 3 signaling with double-stranded RNA.
Science, 320, 2008
|
|
3FHE
 
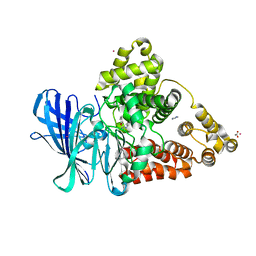 | |
3FH7
 
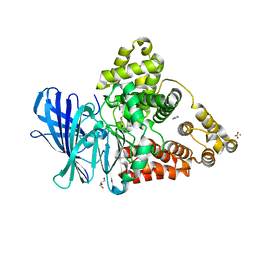 | |
2TRS
 
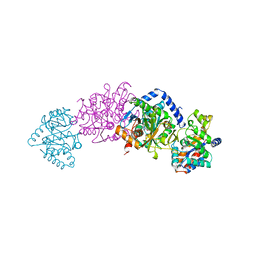 | | CRYSTAL STRUCTURES OF MUTANT (BETAK87T) TRYPTOPHAN SYNTHASE ALPHA2 BETA2 COMPLEX WITH LIGANDS BOUND TO THE ACTIVE SITES OF THE ALPHA AND BETA SUBUNITS REVEAL LIGAND-INDUCED CONFORMATIONAL CHANGES | | Descriptor: | INDOLE-3-PROPANOL PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE, ... | | Authors: | Rhee, S, Parris, K.D, Hyde, C.C, Ahmed, S.A, Miles, E.W, Davies, D.R. | | Deposit date: | 1997-01-07 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes.
Biochemistry, 36, 1997
|
|
2TSY
 
 | | CRYSTAL STRUCTURES OF MUTANT (BETAK87T) TRYPTOPHAN SYNTHASE ALPHA2 BETA2 COMPLEX WITH LIGANDS BOUND TO THE ACTIVE SITES OF THE ALPHA AND BETA SUBUNITS REVEAL LIGAND-INDUCED CONFORMATIONAL CHANGES | | Descriptor: | SN-GLYCEROL-3-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE, ... | | Authors: | Rhee, S, Parris, K.D, Hyde, C.C, Ahmed, S.A, Miles, E.W, Davies, D.R. | | Deposit date: | 1997-01-03 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes.
Biochemistry, 36, 1997
|
|
2TYS
 
 | | CRYSTAL STRUCTURES OF MUTANT (BETAK87T) TRYPTOPHAN SYNTHASE ALPHA2 BETA2 COMPLEX WITH LIGANDS BOUND TO THE ACTIVE SITES OF THE ALPHA AND BETA SUBUNITS REVEAL LIGAND-INDUCED CONFORMATIONAL CHANGES | | Descriptor: | SODIUM ION, TRYPTOPHAN SYNTHASE, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-L-TRYPTOPHANE | | Authors: | Rhee, S, Parris, K.D, Hyde, C.C, Ahmed, S.A, Miles, E.W, Davies, D.R. | | Deposit date: | 1997-01-08 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes.
Biochemistry, 36, 1997
|
|
1TTP
 
 | | TRYPTOPHAN SYNTHASE (E.C.4.2.1.20) IN THE PRESENCE OF CESIUM, ROOM TEMPERATURE | | Descriptor: | CESIUM ION, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN SYNTHASE | | Authors: | Rhee, S, Parris, K, Ahmed, S, Miles, E.W, Davies, D.R. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exchange of K+ or Cs+ for Na+ induces local and long-range changes in the three-dimensional structure of the tryptophan synthase alpha2beta2 complex.
Biochemistry, 35, 1996
|
|
1TTQ
 
 | | TRYPTOPHAN SYNTHASE (E.C.4.2.1.20) IN THE PRESENCE OF POTASSIUM AT ROOM TEMPERATURE | | Descriptor: | POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN SYNTHASE | | Authors: | Rhee, S, Parris, K, Ahmed, S, Miles, E.W, Davies, D.R. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exchange of K+ or Cs+ for Na+ induces local and long-range changes in the three-dimensional structure of the tryptophan synthase alpha2beta2 complex.
Biochemistry, 35, 1996
|
|
1UBS
 
 | | TRYPTOPHAN SYNTHASE (E.C.4.2.1.20) WITH A MUTATION OF LYS 87->THR IN THE B SUBUNIT AND IN THE PRESENCE OF LIGAND L-SERINE | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE, SODIUM ION, ... | | Authors: | Rhee, S, Parris, K, Ahmed, S.A, Miles, E.W, Davies, D.R. | | Deposit date: | 1995-12-14 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes.
Biochemistry, 36, 1997
|
|
1RQD
 
 | | deoxyhypusine synthase holoenzyme in its low ionic strength, high pH crystal form with the inhibitor GC7 bound in the active site | | Descriptor: | 1-GUANIDINIUM-7-AMINOHEPTANE, Deoxyhypusine synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Umland, T.C, Wolff, E.C, Park, M.-H, Davies, D.R. | | Deposit date: | 2003-12-04 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A New Crystal Structure of Deoxyhypusine Synthase Reveals the Configuration of the Active Enzyme and of an Enzyme-NAD-Inhibitor Ternary Complex
J.Biol.Chem., 279, 2004
|
|
1RLZ
 
 | | Deoxyhypusine synthase holoenzyme in its high ionic strength, low pH crystal form | | Descriptor: | Deoxyhypusine synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Umland, T.C, Wolff, E.C, Park, M.-H, Davies, D.R. | | Deposit date: | 2003-11-26 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A New Crystal Structure of Deoxyhypusine Synthase Reveals the Configuration of the Active Enzyme and of an Enzyme-NAD-Inhibitor Ternary Complex
J.Biol.Chem., 279, 2004
|
|
1ROZ
 
 | | Deoxyhypusine synthase holoenzyme in its low ionic strength, high pH crystal form | | Descriptor: | Deoxyhypusine synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Umland, T.C, Wolff, E.C, Park, M.-H, Davies, D.R. | | Deposit date: | 2003-12-02 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A New Crystal Structure of Deoxyhypusine Synthase Reveals the Configuration of the Active Enzyme and of an Enzyme-NAD-Inhibitor Ternary Complex
J.Biol.Chem., 279, 2004
|
|
4NPC
 
 | | Crystal Structure of an Oxidoreductase, Short-Chain Dehydrogenase/Reductase Family Protein from Brucella suis | | Descriptor: | ACETATE ION, Sorbitol dehydrogenase | | Authors: | Dranow, D.M, Davies, D.R, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2013-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of an Oxidoreductase, Short-Chain Dehydrogenase/Reductase Family Protein from Brucella suis
To be Published
|
|
2IFF
 
 | |
1QS4
 
 | | Core domain of HIV-1 integrase complexed with Mg++ and 1-(5-chloroindol-3-yl)-3-hydroxy-3-(2H-tetrazol-5-yl)-propenone | | Descriptor: | 1-(5-CHLOROINDOL-3-YL)-3-HYDROXY-3-(2H-TETRAZOL-5-YL)-PROPENONE, MAGNESIUM ION, PROTEIN (HIV-1 INTEGRASE (E.C.2.7.7.49)) | | Authors: | Goldgur, Y, Craigie, R, Fujiwara, T, Yoshinaga, T, Davies, D.R. | | Deposit date: | 1999-06-25 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1WY4
 
 | | Chicken villin subdomain HP-35, K65(NLE), N68H, pH5.1 | | Descriptor: | IODIDE ION, SODIUM ION, Villin | | Authors: | Chiu, T.K, Kubelka, J, Herbst-Irmer, R, Eaton, W.A, Hofrichter, J, Davies, D.R. | | Deposit date: | 2005-02-04 | | Release date: | 2005-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High-resolution x-ray crystal structures of the villin headpiece subdomain, an ultrafast folding protein.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1WY3
 
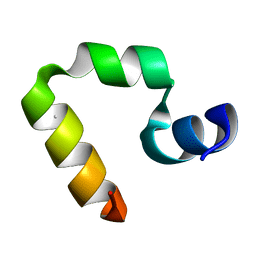 | | Chicken villin subdomain HP-35, K65(NLE), N68H, pH7.0 | | Descriptor: | Villin | | Authors: | Chiu, T.K, Kubelka, J, Herbst-Irmer, R, Eaton, W.A, Hofrichter, J, Davies, D.R. | | Deposit date: | 2005-02-04 | | Release date: | 2005-05-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | High-resolution x-ray crystal structures of the villin headpiece subdomain, an ultrafast folding protein.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1BQL
 
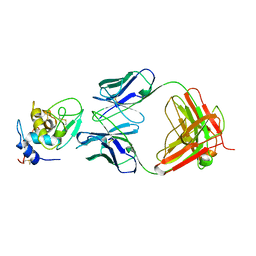 | |
1BIS
 
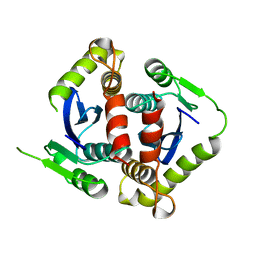 | | HIV-1 INTEGRASE CORE DOMAIN | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-19 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BIZ
 
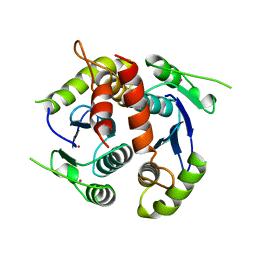 | | HIV-1 INTEGRASE CORE DOMAIN | | Descriptor: | CACODYLATE ION, HIV-1 INTEGRASE | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-21 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1D3R
 
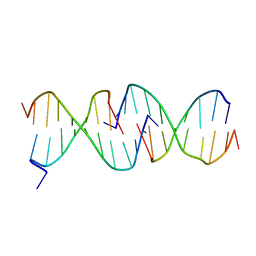 | | CRYSTAL STRUCTURE OF TRIPLEX DNA | | Descriptor: | DNA (5'-D(*CP*(BRU)P*CP*CP*(BRU)P*CP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*CP*GP*GP*AP*G)-3') | | Authors: | Rhee, S, Han, Z.-J, Liu, K, Todd Miles, H.T, Davies, D.R. | | Deposit date: | 1999-09-30 | | Release date: | 2000-01-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a triple helical DNA with a triplex-duplex junction.
Biochemistry, 38, 1999
|
|
1BIU
 
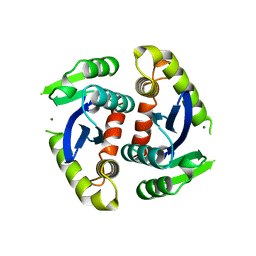 | | HIV-1 INTEGRASE CORE DOMAIN COMPLEXED WITH MG++ | | Descriptor: | HIV-1 INTEGRASE, MAGNESIUM ION | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-19 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BEU
 
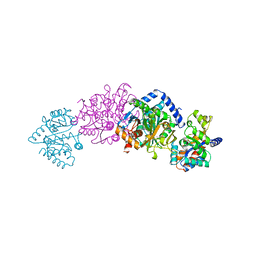 | | TRP SYNTHASE (D60N-IPP-SER) WITH K+ | | Descriptor: | INDOLE-3-PROPANOL PHOSPHATE, POTASSIUM ION, TRYPTOPHAN SYNTHASE, ... | | Authors: | Rhee, S, Mozzarelli, A, Miles, E.W, Davies, D.R. | | Deposit date: | 1998-05-18 | | Release date: | 1998-08-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cryocrystallography and microspectrophotometry of a mutant (alpha D60N) tryptophan synthase alpha 2 beta 2 complex reveals allosteric roles of alpha Asp60.
Biochemistry, 37, 1998
|
|
1CI4
 
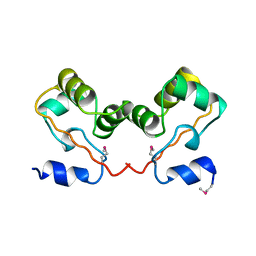 | |
1DHS
 
 | |
