8WGJ
 
 | |
8WGI
 
 | |
3M0C
 
 | |
3SA0
 
 | | Complex of ERK2 with norathyriol | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, norathyriol | | Authors: | Kurinov, I, Malakhova, M. | | Deposit date: | 2011-06-02 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5947 Å) | | Cite: | Norathyriol Suppresses Skin Cancers Induced by Solar Ultraviolet Radiation by Targeting ERK Kinases.
Cancer Res., 72, 2012
|
|
7Y8C
 
 | | Crystal structure of CotA laccase complexed with syringaldehyde | | Descriptor: | 3,5-dimethoxy-4-oxidanyl-benzaldehyde, COPPER (II) ION, Spore coat protein A, ... | | Authors: | Liu, Z.C, Xie, T, Wang, G.G. | | Deposit date: | 2022-06-23 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular insights into substrate promiscuity of CotA laccase catalyzing lignin-phenol derivatives.
Int.J.Biol.Macromol., 256, 2023
|
|
7Y8B
 
 | | Crystal structure of CotA laccase complexed with syringic acid | | Descriptor: | 3,5-dimethoxy-4-oxidanyl-benzoic acid, COPPER (II) ION, Spore coat protein A, ... | | Authors: | Liu, Z.C, Xie, T, Wang, G.G. | | Deposit date: | 2022-06-23 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular insights into substrate promiscuity of CotA laccase catalyzing lignin-phenol derivatives.
Int.J.Biol.Macromol., 256, 2023
|
|
8I54
 
 | | Lb2Cas12a RNA DNA complex | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*GP*TP*GP*CP*TP*TP*TP*A)-3'), Lb2Cas12a, ... | | Authors: | Li, J, Sivaraman, J, Satoru, M. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structures of apo Cas12a and its complex with crRNA and DNA reveal the dynamics of ternary complex formation and target DNA cleavage.
Plos Biol., 21, 2023
|
|
4TXS
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | (4-hydroxyphenyl)acetonitrile, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-07 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TY8
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | 6-methyl-2H-chromen-2-one, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
5PGV
 
 | | CRYSTAL STRUCTURE OF 11BETA-HSD1 DOUBLE MUTANT (L262R, F278E) COMPLEXED WITH 1-(3-HYDROXYAZETIDIN-1-YL)-2-[(2S,5R)-2-(4-FLUOROPHENYL)-5-METHOXYADAMANTAN-2-YL]ETHAN-1-ONE | | Descriptor: | 1-(3-HYDROXYAZETIDIN-1-YL)-2-[(2S,5R)-2-(4-FLUOROPHENYL)-5-METHOXYADAMANTAN-2-YL]ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
5PGW
 
 | | CRYSTAL STRUCTURE OF 11BETA-HSD1 DOUBLE MUTANT (L262R, F278E) COMPLEXED WITH 2-[(1R,3S,5R,7S)-2-[4-(4-FLUOROPHENYL)PHENYL]-6-HYDROXYADAMANTAN-2-YL]-1-(3- HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE | | Descriptor: | 2-[(1R,3S,5R,7S)-2-[4-(4-FLUOROPHENYL)PHENYL]-6-HYDROXYADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
8ZW0
 
 | | Cryo-EM strcuture of Prostaglandin D2 Receptor DP1 activated by PGD2 | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, J, Xu, Y, Wu, C, Xu, H.E. | | Deposit date: | 2024-06-12 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Molecular basis for ligand recognition and receptor activation of the prostaglandin D2 receptor DP1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8ZVZ
 
 | | Cryo-EM strcuture of Prostaglandin D2 Receptor DP1 activated by BW245C | | Descriptor: | 7-{(4S)-3-[(3R)-3-cyclohexyl-3-hydroxypropyl]-2,5-dioxoimidazolidin-4-yl}heptanoic acid, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, J, Xu, Y, Wu, C, Xu, H.E. | | Deposit date: | 2024-06-12 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Molecular basis for ligand recognition and receptor activation of the prostaglandin D2 receptor DP1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8ZTC
 
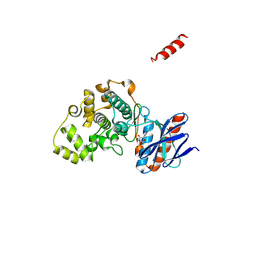 | | Crystal structure of Mps1-AMP complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Mitogen-activated protein kinase MPS1 | | Authors: | Kong, Z, Li, S, Zhang, X, Wang, D, Liu, J. | | Deposit date: | 2024-06-07 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Combinatorial Targeting of Common Docking and ATP Binding Sites on Mps1 MAPK for Management of Pathogenic Fungi.
J.Agric.Food Chem., 72, 2024
|
|
3S5J
 
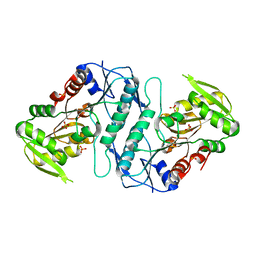 | |
5D1K
 
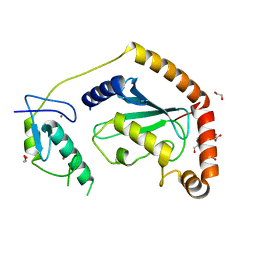 | | Crystal Structure of UbcH5B in Complex with the RING-U5BR Fragment of AO7 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase RNF25, ... | | Authors: | Liang, Y.-H, Li, S, Weissman, A.M, Ji, X. | | Deposit date: | 2015-08-04 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Insights into Ubiquitination from the Unique Clamp-like Binding of the RING E3 AO7 to the E2 UbcH5B.
J.Biol.Chem., 290, 2015
|
|
9M7V
 
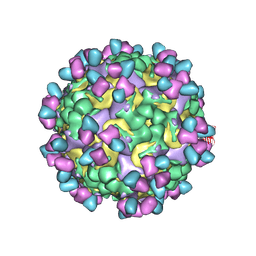 | | Cryo-EM structure of enterovirus A71 mature virion in complex with Fab CT11F9 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Jiang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2025-03-11 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Development of In Vitro Potency Methods to Replace In Vivo Tests for Enterovirus 71 Inactivated Vaccine (Human Diploid Cell-Based/Vero Cell-Based).
Vaccines (Basel), 13, 2025
|
|
5XMA
 
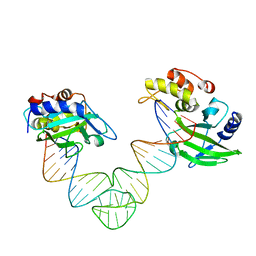 | |
5XM9
 
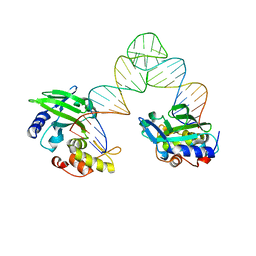 | |
5XWR
 
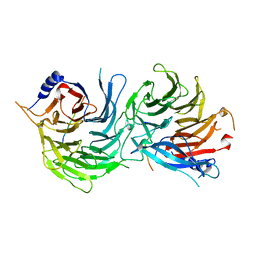 | | Crystal Structure of RBBP4-peptide complex | | Descriptor: | Histone-binding protein RBBP4, MET-SER-ARG-ARG-LYS-GLN-ALA-LYS-PRO-GLN-HIS-ILE | | Authors: | Jobichen, C, Lui, B.H, Daniel, G.T, Sivaraman, J. | | Deposit date: | 2017-06-30 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Targeting cancer addiction for SALL4 by shifting its transcriptome with a pharmacologic peptide.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3JUE
 
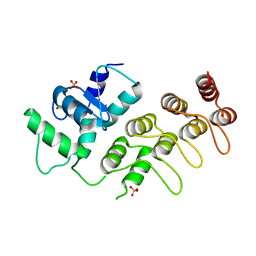 | | Crystal Structure of ArfGAP and ANK repeat domain of ACAP1 | | Descriptor: | ARFGAP with coiled-coil, ANK repeat and PH domain-containing protein 1, SULFATE ION, ... | | Authors: | Pang, X, Zhang, K, Ma, J, Zhou, Q, Sun, F. | | Deposit date: | 2009-09-15 | | Release date: | 2010-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insights into regulated cargo binding by ACAP1 protein
J.Biol.Chem., 287, 2012
|
|
6ZCI
 
 | | Crystal structure of BRD4-BD1 in complex with NVS-BET-1 | | Descriptor: | (4~{R})-4-(4-chlorophenyl)-1-cyclopropyl-5-(1,5-dimethyl-6-oxidanylidene-pyridin-3-yl)-3-methyl-4~{H}-pyrrolo[3,4-c]pyrazol-6-one, Bromodomain-containing protein 4 | | Authors: | Faller, M. | | Deposit date: | 2020-06-11 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.976 Å) | | Cite: | BET bromodomain inhibitors regulate keratinocyte plasticity.
Nat.Chem.Biol., 17, 2021
|
|
6L5M
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
9UWD
 
 | | Cryo-EM structure of inactive-DP1 | | Descriptor: | Prostaglandin D2 receptor,Soluble cytochrome b562 | | Authors: | Xu, J, Xu, Y, Wu, C, Xu, H.E. | | Deposit date: | 2025-05-12 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Molecular basis for ligand recognition and receptor activation of the prostaglandin D2 receptor DP1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
6L5L
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at apo state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
