6SV2
 
 | |
6SUZ
 
 | |
1RT3
 
 | | AZT DRUG RESISTANT HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH 1051U91 | | Descriptor: | 6,11-DIHYDRO-11-ETHYL-6-METHYL-9-NITRO-5H-PYRIDO[2,3-B][1,5]BENZODIAZEPIN-5-ONE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1998-06-29 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 3'-Azido-3'-deoxythymidine drug resistance mutations in HIV-1 reverse transcriptase can induce long range conformational changes.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1RT7
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH UC84 | | Descriptor: | 1-METHYL ETHYL 1-CHLORO-5-[[(5,6DIHYDRO-2-METHYL-1,4-OXATHIIN-3-YL)CARBONYL]AMINO]BENZOATE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptase in complex with carboxanilide derivatives.
Biochemistry, 37, 1998
|
|
1RT5
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH UC10 | | Descriptor: | HIV-1 REVERSE TRANSCRIPTASE, N-[4-CLORO-3-(T-BUTYLOXOME)PHENYL-2-METHYL-3-FURAN-CARBOTHIAMIDE, PHOSPHATE ION | | Authors: | Ren, J, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptase in complex with carboxanilide derivatives.
Biochemistry, 37, 1998
|
|
1RT6
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH UC38 | | Descriptor: | 1-METHYL ETHYL 2-CHLORO-5-[[[(1-METHYLETHOXY)THIOOXO]METHYL]AMINO]-BENZOATE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptase in complex with carboxanilide derivatives.
Biochemistry, 37, 1998
|
|
1RT4
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH UC781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptase in complex with carboxanilide derivatives.
Biochemistry, 37, 1998
|
|
6RST
 
 | | TBK1 in complex with inhibitor compound 24 | | Descriptor: | 1-[4-[(1~{R})-1-[2-[[5-[1-(cyclopropylmethyl)pyrazol-4-yl]-1~{H}-benzimidazol-2-yl]amino]pyridin-4-yl]ethyl]piperazin-1-yl]-3,3,3-tris(fluoranyl)propan-1-one, Serine/threonine-protein kinase TBK1 | | Authors: | Panne, D, Hillig, R.C, Rengachari, S. | | Deposit date: | 2019-05-22 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Discovery of BAY-985, a Highly Selective TBK1/IKK epsilon Inhibitor.
J.Med.Chem., 63, 2020
|
|
6GXB
 
 | | Carbonic Anhydrase CAIX mimic in complex with inhibitor JS13 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, ~{N}-[4-[4-[(4-sulfamoylphenyl)carbamoylamino]phenoxy]butyl]ethanamide | | Authors: | Brynda, J, Rezacova, P, Pospisilova, K. | | Deposit date: | 2018-06-27 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Inhibitor-Polymer Conjugates as a Versatile Tool for Detection and Visualization of Cancer-Associated Carbonic Anhydrase Isoforms
Acs Omega, 4, 2019
|
|
8EST
 
 | | REACTION OF PORCINE PANCREATIC ELASTASE WITH 7-SUBSTITUTED 3-ALKOXY-4-CHLOROISOCOUMARINS: DESIGN OF POTENT INHIBITORS USING THE CRYSTAL STRUCTURE OF THE COMPLEX FORMED WITH 4-CHLORO-3-ETHOXY-7-GUANIDINO-ISOCOUMARIN | | Descriptor: | CALCIUM ION, ETHYL-(2-CARBOXY-4-GUANIDINIUM-PHENYL)-CHLOROACETATE, PORCINE PANCREATIC ELASTASE, ... | | Authors: | Radhakrishnan, R, Powers, J.C, Meyerjunior, E.F. | | Deposit date: | 1990-02-21 | | Release date: | 1992-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Reaction of porcine pancreatic elastase with 7-substituted 3-alkoxy-4-chloroisocoumarins: design of potent inhibitors using the crystal structure of the complex formed with 4-chloro-3-ethoxy-7-guanidinoisocoumarin.
Biochemistry, 29, 1990
|
|
6TSG
 
 | | Crystal structure of Peroxisome proliferator-activated receptor gamma (PPARG) in complex with TETRAC | | Descriptor: | 3,3',5,5'-TETRAIODOTHYROACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Gellrich, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | l-Thyroxin and the Nonclassical Thyroid Hormone TETRAC Are Potent Activators of PPAR gamma.
J.Med.Chem., 63, 2020
|
|
6GXE
 
 | | Carbonic Anhydrase CAIX mimic in complex with inhibitor JS14 | | Descriptor: | 3-[2-[2-[2-[2-(aminomethyloxy)ethoxy]ethoxy]ethoxy]ethoxy]-~{N}-[4-[4-[(4-sulfamoylphenyl)carbamoylamino]phenoxy]butyl]propanamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Brynda, J, Rezacova, P, Pospisilova, K. | | Deposit date: | 2018-06-27 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Inhibitor-Polymer Conjugates as a Versatile Tool for Detection and Visualization of Cancer-Associated Carbonic Anhydrase Isoforms
Acs Omega, 2019
|
|
5ILO
 
 | | Crystal structure of photoreceptor dehydrogenase from Drosophila melanogaster | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Photoreceptor dehydrogenase, isoform C | | Authors: | Hofmann, L, Tsybovsky, Y, Banerjee, S. | | Deposit date: | 2016-03-04 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural Insights into the Drosophila melanogaster Retinol Dehydrogenase, a Member of the Short-Chain Dehydrogenase/Reductase Family.
Biochemistry, 55, 2016
|
|
6HP0
 
 | | Complex of Neuraminidase from H1N1 Influenza Virus in Complex with Oseltamivir Triazol Derivative | | Descriptor: | (3~{R},4~{R},5~{S})-4-acetamido-5-[4-(hydroxymethyl)-1,2,3-triazol-1-yl]-3-pentan-3-yloxy-cyclohexene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J. | | Deposit date: | 2018-09-19 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Investigation of flexibility of neuraminidase 150-loop using tamiflu derivatives in influenza A viruses H1N1 and H5N1.
Bioorg.Med.Chem., 27, 2019
|
|
3KYR
 
 | | Bace-1 in complex with a norstatine type inhibitor | | Descriptor: | 3-[[(2S)-2-[[[(2S)-2-[[(2S)-2-[[(2S)-2-azanyl-3-(1H-1,2,3,4-tetrazol-5-ylcarbonylamino)propanoyl]amino]-3-methyl-butanoyl]amino]-4-methyl-pentanoyl]amino]methyl]-2-hydroxy-4-phenyl-butanoyl]amino]benzoic acid, Beta-secretase 1 | | Authors: | Lindberg, J.D, Borkakoti, N, Derbyshire, D, Nystrom, S. | | Deposit date: | 2009-12-07 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Investigation of a-phenylnorstatine and a-benzylnorstatine as transition state isostere motifs in the search for new BACE-1 inhibiotrs
To be Published
|
|
7R6Z
 
 | | OXA-48 bound by Compound 3.3 | | Descriptor: | 1,2-ETHANEDIOL, 4-amino-5-hydroxynaphthalene-2,7-disulfonic acid, Beta-lactamase, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Sankaran, B, Palzkill, T. | | Deposit date: | 2021-06-24 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
5ILG
 
 | | Crystal structure of photoreceptor dehydrogenase from Drosophila melanogaster | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Hofmann, L, Tsybovsky, Y, Banerjee, S. | | Deposit date: | 2016-03-04 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into the Drosophila melanogaster Retinol Dehydrogenase, a Member of the Short-Chain Dehydrogenase/Reductase Family.
Biochemistry, 55, 2016
|
|
5KES
 
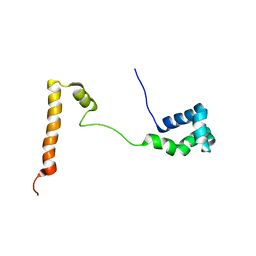 | | Solution structure of the yeast Ddi1 HDD domain | | Descriptor: | DNA damage-inducible protein 1 | | Authors: | Trempe, J.-F, Ratcliffe, C, Veverka, V, Saskova, K, Gehring, K. | | Deposit date: | 2016-06-10 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies of the yeast DNA damage-inducible protein Ddi1 reveal domain architecture of this eukaryotic protein family.
Sci Rep, 6, 2016
|
|
5KL1
 
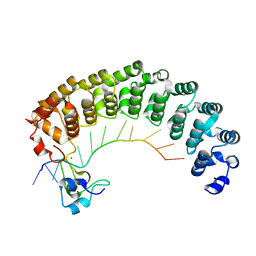 | | Crystal structure of the Pumilio-Nos-hunchback RNA complex | | Descriptor: | Maternal protein pumilio, Protein nanos, RNA (5'-R(*AP*AP*AP*UP*UP*GP*UP*AP*CP*AP*UP*A)-3'), ... | | Authors: | Qiu, C, Hall, T.M.T. | | Deposit date: | 2016-06-23 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.701 Å) | | Cite: | Drosophila Nanos acts as a molecular clamp that modulates the RNA-binding and repression activities of Pumilio.
Elife, 5, 2016
|
|
5KL8
 
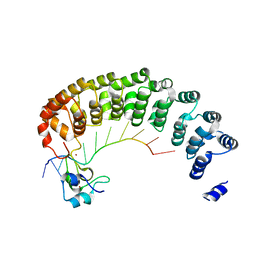 | | Crystal structure of the Pumilio-Nos-CyclinB RNA complex | | Descriptor: | Maternal protein pumilio, Protein nanos, RNA (5'-R(*UP*AP*UP*UP*UP*GP*UP*AP*AP*UP*U)-3'), ... | | Authors: | Qiu, C, Hall, T.M.T. | | Deposit date: | 2016-06-23 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Drosophila Nanos acts as a molecular clamp that modulates the RNA-binding and repression activities of Pumilio.
Elife, 5, 2016
|
|
1SL9
 
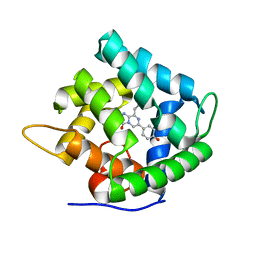 | | Obelin from Obelia longissima | | Descriptor: | C2-HYDROPEROXY-COELENTERAZINE, Obelin | | Authors: | Deng, L, Markova, S, Vysotski, E, Liu, Z.-J, Lee, J, Rose, J, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-05 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Obelin from Obelia longissima
To be Published
|
|
1SL8
 
 | | Calcium-loaded apo-aequorin from Aequorea victoria | | Descriptor: | Aequorin 1, CALCIUM ION | | Authors: | Deng, L, Markova, S.V, Vysotski, E.S, Liu, Z.J, Lee, J, Rose, J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-05 | | Release date: | 2004-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | All three Ca2+-binding loops of photoproteins bind calcium ions: The crystal structures of calcium-loaded apo-aequorin and apo-obelin.
Protein Sci., 14, 2005
|
|
1SL7
 
 | | Crystal structure of calcium-loaded apo-obelin from Obelia longissima | | Descriptor: | CALCIUM ION, Obelin | | Authors: | Deng, L, Markova, S.V, Vysotski, E.S, Liu, Z.J, Lee, J, Rose, J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-05 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | All three Ca2+-binding loops of photoproteins bind calcium ions: The crystal structures of calcium-loaded apo-aequorin and apo-obelin.
Protein Sci., 14, 2005
|
|
8CPH
 
 | | Crystal structure of PPAR gamma (PPARG) in complex with WY-14643 (inactive form) | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
8CPI
 
 | | Crystal structure of PPAR gamma (PPARG) in complex with WY-14643 | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
