5FUL
 
 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 with SAH | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-01-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
FEBS J., 284, 2017
|
|
5FLG
 
 | | Crystal structure of the 6-carboxyhexanoate-CoA ligase (BioW)from Bacillus subtilis in complex with AMPPNP | | Descriptor: | 6-CARBOXYHEXANOATE--COA LIGASE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Moynie, L, Wang, M, Campopiano, D.J, Naismith, J.H. | | Deposit date: | 2015-10-26 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Using the pimeloyl-CoA synthetase adenylation fold to synthesize fatty acid thioesters.
Nat. Chem. Biol., 13, 2017
|
|
4YG0
 
 | | Structural basis of glycan recognition in neonate-specific rotaviruses | | Descriptor: | Outer capsid protein VP4, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hu, L, Prasad, B.V.V. | | Deposit date: | 2015-02-25 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.285 Å) | | Cite: | Structural basis of glycan specificity in neonate-specific bovine-human reassortant rotavirus.
Nat Commun, 6, 2015
|
|
4YL9
 
 | | Crystal Structure of wild-type of hsp14.1 from Sulfolobus solfatataricus P2 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Heat shock protein Hsp20 | | Authors: | Liu, L, Chen, J.Y, Yun, C.H. | | Deposit date: | 2015-03-05 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Active-State Structures of a Small Heat-Shock Protein Revealed a Molecular Switch for Chaperone Function
Structure, 23, 2015
|
|
5G0Q
 
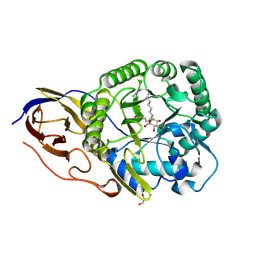 | | beta-glucuronidase with an activity-based probe (N-alkyl cyclophellitol aziridine) bound | | Descriptor: | (1R,2S,3R,4S,5S,6R)-7-(8-AZIDOOCTYL)-3,4,5-TRIHYDROXY-, BETA-GLUCURONIDASE, GLYCEROL | | Authors: | Jin, Y, Wu, L, Jiang, J.B, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2016-03-22 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
7MPH
 
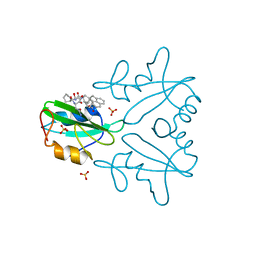 | | GRB2 SH2 Domain with Compound 7 | | Descriptor: | (4-{(10R,11E,14S,18S)-18-(2-amino-2-oxoethyl)-14-[(naphthalen-1-yl)methyl]-8,17,20-trioxo-7,16,19-triazaspiro[5.14]icos-11-en-10-yl}phenyl)acetic acid, 1,2-ETHANEDIOL, Growth factor receptor-bound protein 2, ... | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and structural characterization of a monocarboxylic inhibitor for GRB2 SH2 domain.
Bioorg.Med.Chem.Lett., 51, 2021
|
|
5G04
 
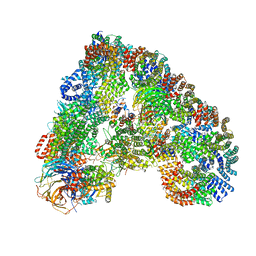 | | Structure of the human APC-Cdc20-Hsl1 complex | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Zhang, S, Chang, L, Alfieri, C, Zhang, Z, Yang, J, Maslen, S, Skehel, M, Barford, D. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular Mechanism of Apc/C Activation by Mitotic Phosphorylation.
Nature, 533, 2016
|
|
4YO3
 
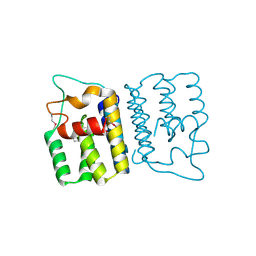 | | Enteroaggregative Escherichia Coli TssA N-terminal fragment | | Descriptor: | TssA | | Authors: | Durand, E, Zoued, A, Spinelli, S, Douzi, B, Brunet, Y.R, Bebeacua, C, Legrand, P, Journet, L, Mignot, T, Cambillau, C, Cascales, E. | | Deposit date: | 2015-03-11 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Priming and polymerization of a bacterial contractile tail structure.
Nature, 531, 2016
|
|
5FSU
 
 | |
5FPQ
 
 | | Structure of Homo sapiens acetylcholinesterase phosphonylated by sarin. | | Descriptor: | ACETYLCHOLINESTERASE, PENTAETHYLENE GLYCOL | | Authors: | Allgardsson, A, Berg, L, Akfur, C, Hornberg, A, Worek, F, Linusson, A, Ekstrom, F. | | Deposit date: | 2015-12-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Prereaction Complex between the Nerve Agent Sarin, its Biological Target Acetylcholinesterase, and the Antidote Hi-6.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4YPR
 
 | | Crystal Structure of D144N MutY bound to its anti-substrate | | Descriptor: | A/G-specific adenine glycosylase, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), DNA (5'-D(*T*GP*TP*CP*CP*AP*CP*GP*TP*CP*T)-3'), ... | | Authors: | Wang, L, Lee, S, Verdine, G.L. | | Deposit date: | 2015-03-13 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural Basis for Avoidance of Promutagenic DNA Repair by MutY Adenine DNA Glycosylase.
J.Biol.Chem., 290, 2015
|
|
7MQJ
 
 | | Dhr1 Helicase Core | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Probable ATP-dependent RNA helicase DHR1 | | Authors: | Miller, L, Chaker-Margot, M, Klinge, S. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Nucleolar maturation of the human small subunit processome.
Science, 373, 2021
|
|
5FOK
 
 | |
5G0H
 
 | | Crystal structure of Danio rerio HDAC6 CD2 in complex with (S)- trichostatin A | | Descriptor: | 1,2-ETHANEDIOL, HDAC6, POTASSIUM ION, ... | | Authors: | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
3OJT
 
 | | Structure of native Fe-containing Homoprotocatechuate 2,3-Dioxygenase at 1.70 Ang resolution | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (II) ION, ... | | Authors: | Fielding, A.J, Kovaleva, E.G, Farquhar, E.R, Lipscomb, J.D, Que Jr, L. | | Deposit date: | 2010-08-23 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A hyperactive cobalt-substituted extradiol-cleaving catechol dioxygenase.
J.Biol.Inorg.Chem., 16, 2011
|
|
5G0F
 
 | | Crystal structure of Danio rerio HDAC6 ZnF-UBP domain | | Descriptor: | GLYCINE, HDAC6, NICKEL (II) ION, ... | | Authors: | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
4Y7S
 
 | | Crystal Structure of the CFEM protein Csa2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, HEME B/C, ... | | Authors: | Dvir, H, Weissman, Z, Nasser, L, Hiya, D, Kornitzer, D. | | Deposit date: | 2015-02-16 | | Release date: | 2016-08-03 | | Last modified: | 2016-10-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of haem-iron acquisition by fungal pathogens.
Nat Microbiol, 1, 2016
|
|
5FRH
 
 | | Solution structure of oxidised RsrA | | Descriptor: | ANTI-SIGMA FACTOR RSRA | | Authors: | Zdanowski, K, Pecqueur, L, Werner, J, Potts, J.R, Kleanthous, C. | | Deposit date: | 2015-12-17 | | Release date: | 2016-08-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Anti-Sigma Factor Rsra Responds to Oxidative Stress by Reburying its Hydrophobic Core.
Nat.Commun., 7, 2016
|
|
5FSZ
 
 | |
5FJN
 
 | | Structure of L-Amino acid deaminase from Proteus myxofaciens in complex with anthranilate | | Descriptor: | 2-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID DEAMINASE | | Authors: | Motta, P, Molla, G, Pollegioni, L, Nardini, M. | | Deposit date: | 2015-10-11 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Function Relationships in L-Amino Acid Deaminase, a Flavoprotein Belonging to a Novel Class of Biotechnologically Relevant Enzymes
J.Biol.Chem., 291, 2016
|
|
5FJM
 
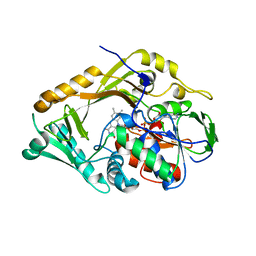 | | Structure of L-Amino acid deaminase from Proteus myxofaciens | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID DEAMINASE | | Authors: | Motta, P, Molla, G, Pollegioni, L, Nardini, M. | | Deposit date: | 2015-10-11 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Function Relationships in L-Amino Acid Deaminase, a Flavoprotein Belonging to a Novel Class of Biotechnologically Relevant Enzymes
J.Biol.Chem., 291, 2016
|
|
5FWD
 
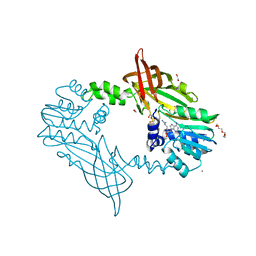 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 with CP2 | | Descriptor: | 1,2-ETHANEDIOL, 9-(7-{[amino(iminio)methyl]amino}-5,6,7-trideoxy-beta-D-ribo-heptofuranosyl)-9H-purin-6-amine, CALCIUM ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-02-12 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Studies of Protein Arginine Methyltransferase 2 Reveal its Interactions with Potential Substrates and Inhibitors.
FEBS J., 284, 2017
|
|
5FQO
 
 | |
5FK1
 
 | |
5FKH
 
 | |
