6BWW
 
 | |
6C51
 
 | | Cross-alpha Amyloid-like Structure alphaAmL | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cross-alpha Amyloid-like Structure alphaAmL, PHOSPHATE ION | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
7O5B
 
 | | Cryo-EM structure of a Bacillus subtilis MifM-stalled ribosome-nascent chain complex with (p)ppGpp-SRP bound | | Descriptor: | 16S rRNA (1533-MER), 23S rRNA (2887-MER), 30S ribosomal protein S10, ... | | Authors: | Kratzat, H, Czech, L, Berninghausen, O, Bange, G, Beckmann, R. | | Deposit date: | 2021-04-08 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Inhibition of SRP-dependent protein secretion by the bacterial alarmone (p)ppGpp.
Nat Commun, 13, 2022
|
|
3F0A
 
 | | Structure of a putative n-acetyltransferase (ta0374) in complex with acetyl-coa from thermoplasma acidophilum | | Descriptor: | ACETYL COENZYME *A, CHLORIDE ION, N-ACETYLTRANSFERASE, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-24 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
3F2W
 
 | |
3F81
 
 | | Interaction of VHR with SA3 | | Descriptor: | 2-[(5~{E})-5-[[3-[4-(2-fluoranylphenoxy)phenyl]-1-phenyl-pyrazol-4-yl]methylidene]-4-oxidanylidene-2-sulfanylidene-1,3-thiazolidin-3-yl]ethanesulfonic acid, Dual specificity protein phosphatase 3 | | Authors: | Wu, S, Mutelin, T, Tautz, L. | | Deposit date: | 2008-11-11 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multidentate small-molecule inhibitors of vaccinia H1-related (VHR) phosphatase decrease proliferation of cervix cancer cells.
J.Med.Chem., 52, 2009
|
|
6BV3
 
 | | Crystal structure of porcine aminopeptidase-N with Leucine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Lin, Y.-L, Li, F. | | Deposit date: | 2017-12-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Rational Design of Therapeutic Peptides for Aminopeptidase N using a Substrate-Based Approach.
Sci Rep, 7, 2017
|
|
3F9G
 
 | |
6C32
 
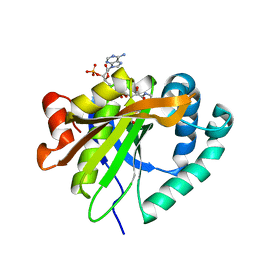 | | Mycobacterium smegmatis RimJ with AcCoA | | Descriptor: | ACETYL COENZYME *A, Acetyltransferase, GNAT family protein | | Authors: | Favrot, L, Hegde, S.S, Blanchard, J.S. | | Deposit date: | 2018-01-09 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Structural Characterization of Mycobacterium smegmatis RimJ, an N-acetyltransferase protein
To Be Published
|
|
6BW4
 
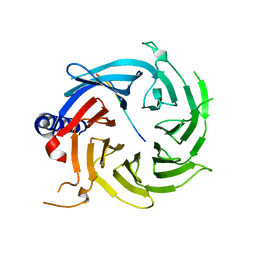 | | Crystal structure of RBBP4 in complex with PRDM16 N-terminal peptide | | Descriptor: | Histone-binding protein RBBP4, PR domain zinc finger protein 16, UNKNOWN ATOM OR ION | | Authors: | Ivanochko, D, Halabelian, L, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-14 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex.
Nucleic Acids Res., 47, 2019
|
|
7NSG
 
 | | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with HIP-B | | Descriptor: | (+)-3-Hydroxy-4,5,6,6a-tetrahydro-3aH-pyrrolo[3,4-d]isoxazole-6-carboxylic acid, (-)-3-Hydroxy-4,5,6,6a-tetrahydro-3aH-pyrrolo[3,4-d]isoxazole-6-carboxylic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Baronina, A, Pike, A.C.W, Yu, X, Dong, Y.Y, Shintre, C.A, Tessitore, A, Chu, A, Rotty, B, Venkaya, S, Mukhopadhyay, S.M.M, Borkowska, O, Chalk, R, Shrestha, L, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Han, S, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-05 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with HIP-B
TO BE PUBLISHED
|
|
6BWM
 
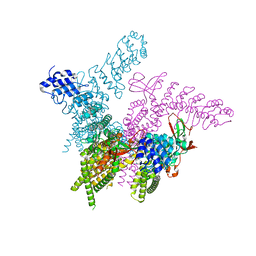 | | Crystal structure of the TRPV2 ion channel | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Zubcevic, L, Le, S, Yang, H, Lee, S.Y. | | Deposit date: | 2017-12-15 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Conformational plasticity in the selectivity filter of the TRPV2 ion channel.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3FCC
 
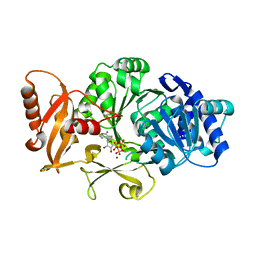 | | CRYSTAL STRUCTURE OF DLTA PROTEIN IN COMPLEX WITH ATP and MAGNESIUM | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-alanine--poly(phosphoribitol) ligase subunit 1, MAGNESIUM ION | | Authors: | Osman, K.T, Du, L, He, Y, Luo, Y. | | Deposit date: | 2008-11-21 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure of Bacillus cereus D-alanyl carrier protein ligase (DltA) in complex with ATP.
J.Mol.Biol., 388, 2009
|
|
7NUE
 
 | | Crystal structure of mouse PRMT6 in complex with inhibitor EML736 | | Descriptor: | Protein arginine N-methyltransferase 6, methyl 6-[5-[[~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]carbamimidoyl]amino]pentylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
3FEH
 
 | | Crystal structure of full length centaurin alpha-1 | | Descriptor: | Centaurin-alpha-1, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Shen, L, Tong, Y, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-29 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phosphorylation-independent dual-site binding of the FHA domain of KIF13 mediates phosphoinositide transport via centaurin {alpha}1.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6C5N
 
 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase with hydroxyoxamate inhibitor 1 | | Descriptor: | (cyclopentylamino)(oxo)acetic acid, IMIDAZOLE, Ketol-acid reductoisomerase (NADP(+)), ... | | Authors: | Kandale, A, Patel, K.M, Zheng, S, You, L, Guddat, L.W, Schenk, G, Schembri, M.A, McGeary, R.P. | | Deposit date: | 2018-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | Design, synthesis, in vitro activity and crystallisation of novel N-isopropyl-N-hydroxyoxamate derivatives as ketol-acid reductosiomerase (KARI) inhibitor
To Be Published
|
|
7NUD
 
 | | Crystal structure of mouse PRMT6 in complex with inhibitor EML734 | | Descriptor: | Protein arginine N-methyltransferase 6, methyl 6-[3-[[~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]carbamimidoyl]amino]propylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
6C7A
 
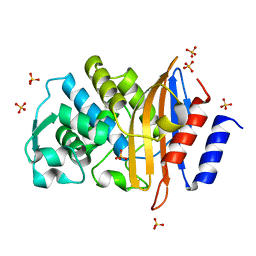 | | Conformational Changes in a Class A Beta lactamase that Prime it for Catalysis | | Descriptor: | Beta-lactamase Toho-1, SULFATE ION | | Authors: | Coates, L, Langan, P.S, Vandavasi, V.G, Cooper, S.J, Weiss, K.L, Ginell, S.L, Parks, J.M. | | Deposit date: | 2018-01-22 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Substrate Binding Induces Conformational Changes in a Class A Beta-lactamase That Prime It for Catalysis
Acs Catalysis, 8, 2018
|
|
3FFC
 
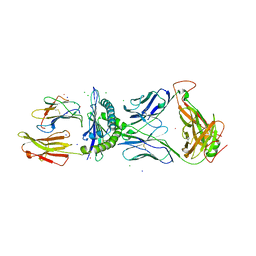 | | Crystal Structure of CF34 TCR in complex with HLA-B8/FLR | | Descriptor: | Beta-2-microglobulin, CADMIUM ION, CF34 alpha chain, ... | | Authors: | Gras, S, Burrows, S.R, Kjer-Nielsen, L, Clements, C.S, Liu, Y.C, Sullivan, L.C, Brooks, A.G, Purcell, A.W, McCluskey, J, Rossjohn, J. | | Deposit date: | 2008-12-03 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The shaping of T cell receptor recognition by self-tolerance.
Immunity, 30, 2009
|
|
6C0M
 
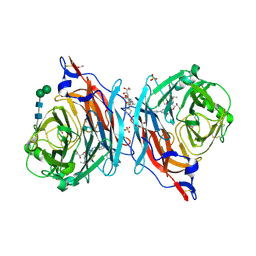 | | The synthesis, biological evaluation and structural insights of unsaturated 3-N-substituted sialic acids as probes of human parainfluenza virus-3 haemagglutinin-neuraminidase | | Descriptor: | 1,2-ETHANEDIOL, 2,6-anhydro-3,5-dideoxy-5-[(2-methylpropanoyl)amino]-3-(4-phenyl-1H-1,2,3-triazol-1-yl)-D-glycero-D-galacto-non-2-enoni c acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dirr, L, Ve, T, von Itzstein, M. | | Deposit date: | 2018-01-01 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Insights into Human Parainfluenza Virus 3 Hemagglutinin-Neuraminidase Using Unsaturated 3- N-Substituted Sialic Acids as Probes.
ACS Chem. Biol., 13, 2018
|
|
7O0Y
 
 | | ABC transporter NosDFY, nucleotide-free in GDN | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O15
 
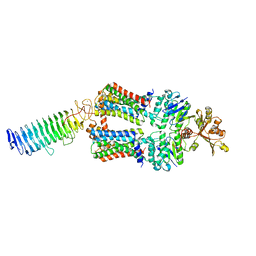 | | ABC transporter NosDFY, nucleotide-free in lipid nanodisc, R-domain 2 | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O16
 
 | | ABC transporter NosDFY, nucleotide-free in lipid nanodisc, R-domain 3 | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
6C4O
 
 | |
7O14
 
 | | ABC transporter NosDFY, nucleotide-free in lipid nanodisc, R-domain 1 | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
