8ZIH
 
 | | Crystal structure of the methyltransferase PsiM in complex with SAH | | Descriptor: | Psilocybin synthase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Meng, C.Y, Wen, Y, Guo, W.T, Wu, B.X. | | Deposit date: | 2024-05-14 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural basis for psilocybin biosynthesis.
Nat Commun, 16, 2025
|
|
8ZIG
 
 | | Crystal structure of the methyltransferase PsiM in complex with s-adenosylmethionine (SAM) | | Descriptor: | Psilocybin synthase, S-ADENOSYLMETHIONINE | | Authors: | Meng, C.Y, Wen, Y, Guo, W.T, Wu, B.X. | | Deposit date: | 2024-05-13 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis for psilocybin biosynthesis.
Nat Commun, 16, 2025
|
|
8ZIC
 
 | | Structure complex of 4-hydroxytryptamine kinase PsiK complexed with Mg2+ and Tryptamine | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANAMINE, 4-hydroxytryptamine kinase, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Meng, C.Y, Wen, Y, Guo, W.T, Wu, B.X. | | Deposit date: | 2024-05-13 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural basis for psilocybin biosynthesis.
Nat Commun, 16, 2025
|
|
8ZII
 
 | | Crystal structure of the methyltransferase PsiM in complex with SAH and Norbaeocystin | | Descriptor: | Norbaeocystin, Psilocybin synthase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Meng, C.Y, Wen, Y, Guo, W.T, Wu, B.X. | | Deposit date: | 2024-05-14 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for psilocybin biosynthesis.
Nat Commun, 16, 2025
|
|
8ZIA
 
 | | The L-tryptophan specific decarboxylase PsiD(50-439) in complex with tryptamine | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANAMINE, L-tryptophan decarboxylase | | Authors: | Meng, C.Y, Wen, Y, Guo, W.T, Wu, B.X. | | Deposit date: | 2024-05-13 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for psilocybin biosynthesis.
Nat Commun, 16, 2025
|
|
8ZIM
 
 | | Crystal structure of the methyltransferase PsiM in complex with SAH and Baeocystin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Baeocystin, GLYCEROL, ... | | Authors: | Meng, C.Y, Wen, Y, Guo, W.T, Wu, B.X. | | Deposit date: | 2024-05-14 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for psilocybin biosynthesis.
Nat Commun, 16, 2025
|
|
8ZIE
 
 | |
7XR6
 
 | | Structure of human excitatory amino acid transporter 2 (EAAT2) in complex with WAY-213613 | | Descriptor: | (2S)-2-azanyl-4-[[4-[2-bromanyl-4,5-bis(fluoranyl)phenoxy]phenyl]amino]-4-oxidanylidene-butanoic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Zhao, Y, Zhang, Z. | | Deposit date: | 2022-05-09 | | Release date: | 2022-06-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of ligand binding modes of human EAAT2.
Nat Commun, 13, 2022
|
|
7XR4
 
 | |
7YEY
 
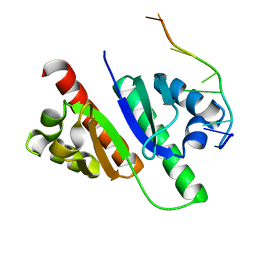 | | Structure of MmIGF2BP3-KH12 in complex with 8-mer RNA | | Descriptor: | Insulin-like growth factor 2 mRNA-binding protein 3, RNA (5'-R(*CP*AP*CP*GP*GP*CP*AP*C)-3') | | Authors: | Li, X.J, Wu, B.X. | | Deposit date: | 2022-07-06 | | Release date: | 2023-07-12 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the RNA binding properties of mouse IGF2BP3.
Structure, 2025
|
|
7YEW
 
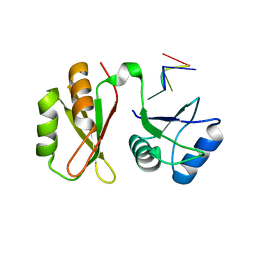 | | Structure of MmIGF2BP3 in complex with 7-mer RNA | | Descriptor: | Insulin-like growth factor 2 mRNA-binding protein 3, RNA (5'-R(P*CP*AP*CP*AP*CP*AP*C)-3') | | Authors: | Li, X.J, Wu, B.X. | | Deposit date: | 2022-07-06 | | Release date: | 2023-07-12 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the RNA binding properties of mouse IGF2BP3.
Structure, 2025
|
|
7YEX
 
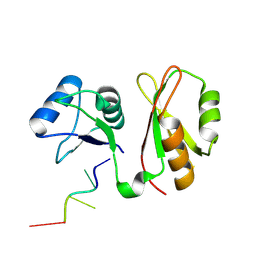 | | Structure of MmIGF2BP3 in complex with 8-mer RNA | | Descriptor: | Insulin-like growth factor 2 mRNA-binding protein 3, RNA (5'-R(P*CP*AP*CP*AP*CP*AP*CP*A)-3') | | Authors: | Li, X.J, Wu, B.X. | | Deposit date: | 2022-07-06 | | Release date: | 2023-07-12 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the RNA binding properties of mouse IGF2BP3.
Structure, 2025
|
|
7JHI
 
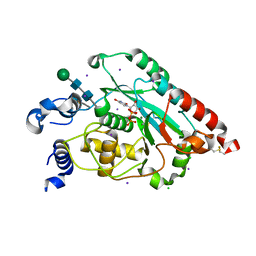 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 iodide-derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
7WJV
 
 | | Crystal structure of human liver FBPase complexed with an covalent inhibitor | | Descriptor: | 1,2-BENZISOTHIAZOL-3(2H)-ONE 1,1-DIOXIDE, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Cao, H, Huang, Y, Ren, Y, Wan, J. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.724 Å) | | Cite: | N -Acylamino Saccharin as an Emerging Cysteine-Directed Covalent Warhead and Its Application in the Identification of Novel FBPase Inhibitors toward Glucose Reduction.
J.Med.Chem., 65, 2022
|
|
7JHK
 
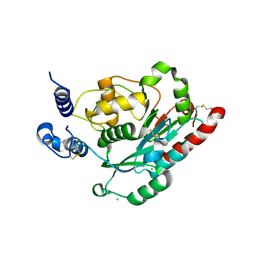 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 in unliganded form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3436 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
7JHM
 
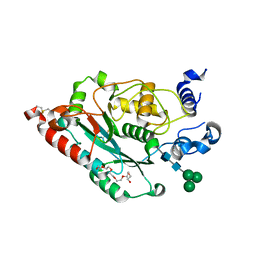 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with N-acetyl-lactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEXAETHYLENE GLYCOL, N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase 2, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
7JHO
 
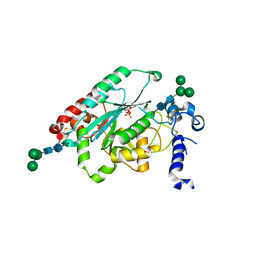 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-21 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
7JHN
 
 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with UDP and trisaccharide GlcNAc-beta1-3Gal-beta1-4GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-21 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
7JHL
 
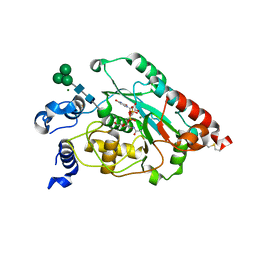 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with UDP-N-acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
7X7S
 
 | | Solution structure of human adenylate kinase 1 (hAK1) | | Descriptor: | Adenylate kinase isoenzyme 1 | | Authors: | Zhang, H. | | Deposit date: | 2022-03-10 | | Release date: | 2022-05-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | ADP-Induced Conformational Transition of Human Adenylate Kinase 1 Is Triggered by Suppressing Internal Motion of alpha 3 alpha 4 and alpha 7 alpha 8 Fragments on the ps-ns Timescale.
Biomolecules, 12, 2022
|
|
7CUJ
 
 | | Crystal structure of fission yeast Ccq1 and Tpz1 | | Descriptor: | Coiled-coil quantitatively-enriched protein 1, Protection of telomeres protein tpz1 | | Authors: | Sun, H, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2020-08-23 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into Pot1-ssDNA, Pot1-Tpz1 and Tpz1-Ccq1 Interactions within fission yeast shelterin complex.
Plos Genet., 18, 2022
|
|
7CUI
 
 | | Crystal structure of fission yeast Pot1 and Tpz1 | | Descriptor: | Protection of telomeres protein 1, Protection of telomeres protein tpz1, SULFATE ION | | Authors: | Sun, H, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2020-08-23 | | Release date: | 2021-08-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into Pot1-ssDNA, Pot1-Tpz1 and Tpz1-Ccq1 Interactions within fission yeast shelterin complex.
Plos Genet., 18, 2022
|
|
7CUH
 
 | | Crystal structure of fission yeast Pot1 and ssDNA | | Descriptor: | Protection of telomeres protein 1, Telomere single-strand DNA | | Authors: | Sun, H, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2020-08-23 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into Pot1-ssDNA, Pot1-Tpz1 and Tpz1-Ccq1 Interactions within fission yeast shelterin complex.
Plos Genet., 18, 2022
|
|
7WD2
 
 | | Crystal structure of S43 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Q.H, Gao, G.F, Qi, J.X, Su, C, Liu, H.H, Wu, L.L. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Two pan-SARS-CoV-2 nanobodies and their multivalent derivatives effectively prevent Omicron infections in mice.
Cell Rep Med, 4, 2023
|
|
7WD1
 
 | | Crystal structure of R14 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, R14, Spike protein S1, ... | | Authors: | Wang, Q.H, Gao, G.F, Qi, J.X, Su, C, Liu, H.H, Wu, L.L. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two pan-SARS-CoV-2 nanobodies and their multivalent derivatives effectively prevent Omicron infections in mice.
Cell Rep Med, 4, 2023
|
|
