5UJO
 
 | |
5UL8
 
 | |
3FDF
 
 | | Crystal structure of the serine phosphatase of RNA polymerase II CTD (SSU72 superfamily) from Drosophila melanogaster. Orthorhombic crystal form. Northeast Structural Genomics Consortium target FR253. | | Descriptor: | FR253 | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Forouhar, F, Chinag, Y, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-11-25 | | Release date: | 2009-01-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Orthorombic crystal structure of serine phosphatase of rna polymerase ii ctd from fly drosofila melanogaster. northeast structural genomics consortium target fr253.
To be Published
|
|
3FMV
 
 | | Crystal structure of the serine phosphatase of RNA polymerase II CTD (SSU72 superfamily) from Drosophila melanogaster. Monoclinic crystal form. Northeast Structural Genomics Consortium target FR253. | | Descriptor: | Serine phosphatase of RNA polymerase II CTD | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Forouhar, F, Chinag, Y, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-22 | | Release date: | 2009-01-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal structure of the serine phosphatase of RNA polymerase II CTD (SSU72 superfamily) from Drosophila melanogaster. Monoclinic crystal form. Northeast Structural Genomics Consortium target FR253.
To be Published
|
|
3DW8
 
 | | Structure of a Protein Phosphatase 2A Holoenzyme with B55 subunit | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Xu, Y, Chen, Y, Zhang, P, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2008-07-21 | | Release date: | 2008-10-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of a protein phosphatase 2A holoenzyme: insights into B55-mediated Tau dephosphorylation.
Mol.Cell, 31, 2008
|
|
6LQZ
 
 | | Solution structure of Taf14ET-Sth1EBMC | | Descriptor: | Nuclear protein STH1/NPS1, Transcription initiation factor TFIID subunit 14 | | Authors: | Wu, B, Chen, G, Chen, Y. | | Deposit date: | 2020-01-15 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Taf14 recognizes a common motif in transcriptional machineries and facilitates their clustering by phase separation.
Nat Commun, 11, 2020
|
|
7KX5
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with noncovalent inhibitor Jun8-76-3A | | Descriptor: | 3C-like proteinase, GLYCEROL, N-([1,1'-biphenyl]-4-yl)-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]furan-2-carboxamide | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2020-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Di- and Trihaloacetamides as Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity.
J.Am.Chem.Soc., 143, 2021
|
|
6NEB
 
 | | MYC Promoter G-Quadruplex with 1:6:1 loop length | | Descriptor: | DNA (27-MER) | | Authors: | Dickerhoff, J, Onel, B, Chen, L, Chen, Y, Yang, D. | | Deposit date: | 2018-12-17 | | Release date: | 2019-02-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a MYC Promoter G-Quadruplex with 1:6:1 Loop Length.
Acs Omega, 4, 2019
|
|
3GX8
 
 | | Structural and biochemical characterization of yeast monothiol glutaredoxin Grx5 | | Descriptor: | Monothiol glutaredoxin-5, mitochondrial, SULFATE ION | | Authors: | Wang, Y, He, Y.X, Yu, J, Xiong, Y, Chen, Y, Zhou, C.Z. | | Deposit date: | 2009-04-01 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | Structural and biochemical characterization of yeast monothiol glutaredoxin Grx5
To be Published
|
|
2QIK
 
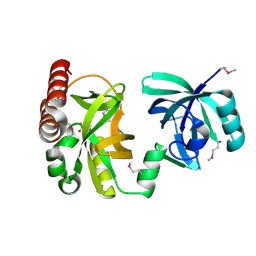 | | Crystal structure of YkqA from Bacillus subtilis. Northeast Structural Genomics Target SR631 | | Descriptor: | CITRIC ACID, UPF0131 protein ykqA | | Authors: | Benach, J, Chen, Y, Forouhar, F, Seetharaman, J, Baran, M.C, Cunningham, K, Ma, L.-C, Owens, L, Chen, C.X, Rong, X, Janjua, H, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-07-05 | | Release date: | 2007-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of YkqA from Bacillus subtilis.
To be Published
|
|
8HLM
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 2) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Guo, M, Wang, H, Wei, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.522 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8HLL
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 1) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Wei, H, Guo, M, Wang, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8HLN
 
 | | Crystal structure of p53/BCL2 fusion complex(complex3) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Guo, M, Wei, H, Wang, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
7K9A
 
 | | Crystal Structure of P. aeruginosa LpxC with N-Hydroxyformamide inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-hydroxy-N-[(1R)-2-{5-[(4-{[2-(hydroxymethyl)-1H-imidazol-1-yl]methyl}phenyl)ethynyl]-1H-benzotriazol-1-yl}-1-(methylsulfanyl)ethyl]formamide, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2020-09-29 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-Hydroxyformamide LpxC inhibitors, their in vivo efficacy in a mouse Escherichia coli infection model, and their safety in a rat hemodynamic assay.
Bioorg.Med.Chem., 28, 2020
|
|
7RPK
 
 | | Cryo-EM structure of murine Dispatched in complex with Sonic hedgehog | | Descriptor: | (2S)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
3J9I
 
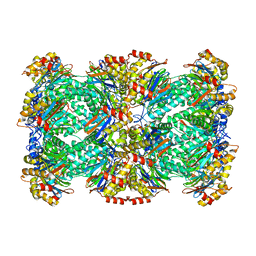 | | Thermoplasma acidophilum 20S proteasome | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Li, X, Mooney, P, Zheng, S, Booth, C, Braunfeld, M.B, Gubbens, S, Agard, D.A, Cheng, Y. | | Deposit date: | 2015-02-02 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM.
Nat.Methods, 10, 2013
|
|
7RPH
 
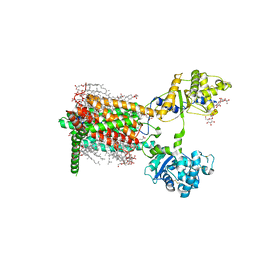 | | Cryo-EM structure of murine Dispatched 'R' conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
7RPJ
 
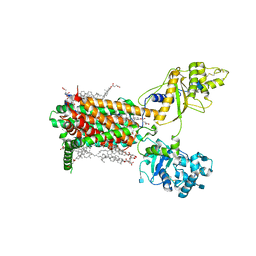 | | Cryo-EM structure of murine Dispatched NNN mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Protein dispatched homolog 1 | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
7RPI
 
 | | Cryo-EM structure of murine Dispatched 'T' conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
2NPP
 
 | | Structure of the Protein Phosphatase 2A Holoenzyme | | Descriptor: | MANGANESE (II) ION, Protein Phosphatase 2, regulatory subunit A (PR 65), ... | | Authors: | Xu, Y, Chen, Y, Xing, Y, Chao, Y, Shi, Y. | | Deposit date: | 2006-10-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the protein phosphatase 2A holoenzyme
Cell(Cambridge,Mass.), 127, 2006
|
|
6AKO
 
 | | Crystal Structure of FOXC2 DBD Bound to DBE2 DNA | | Descriptor: | DNA (5'-D(CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(TP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), Forkhead box protein C2, ... | | Authors: | Chen, X, Wei, H, Li, J, Liang, X, Dai, S, Jiang, L, Guo, M, Chen, Y. | | Deposit date: | 2018-09-03 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Structural basis for DNA recognition by FOXC2.
Nucleic Acids Res., 47, 2019
|
|
6AKP
 
 | | Crystal Structural of FOXC2 DNA binding domain bound to PC promoter | | Descriptor: | DNA (5'-D(AP*CP*AP*CP*AP*AP*AP*TP*AP*TP*TP*TP*GP*TP*GP*T)-3'), Forkhead box protein C2, MAGNESIUM ION | | Authors: | Chen, X, Wei, H, Li, J, Liang, X, Dai, S, Jiang, L, Guo, M, Chen, Y. | | Deposit date: | 2018-09-03 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.323 Å) | | Cite: | Structural basis for DNA recognition by FOXC2.
Nucleic Acids Res., 47, 2019
|
|
2NYL
 
 | | Crystal structure of Protein Phosphatase 2A (PP2A) holoenzyme with the catalytic subunit carboxyl terminus truncated | | Descriptor: | MANGANESE (II) ION, Protein phosphatase 2, regulatory subunit A (PR 65), ... | | Authors: | Xing, Y, Xu, Y, Chen, Y, Chao, Y, Lin, Z, Shi, Y. | | Deposit date: | 2006-11-20 | | Release date: | 2006-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the Protein Phosphatase 2A Holoenzyme.
Cell(Cambridge,Mass.), 127, 2006
|
|
5U4J
 
 | | Structural Basis of Co-translational Quality Control by ArfA and RF2 Bound to Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S12, ... | | Authors: | Zeng, F, Chen, Y, Remis, J, Shekhar, M, Phillips, J.C, Tajkhorshid, E, Jin, H. | | Deposit date: | 2016-12-04 | | Release date: | 2017-01-11 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of co-translational quality control by ArfA and RF2 bound to ribosome.
Nature, 541, 2017
|
|
5TOP
 
 | |
