5H19
 
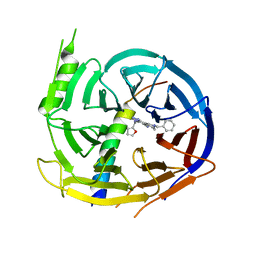 | | EED in complex with PRC2 allosteric inhibitor EED162 | | Descriptor: | 5-(furan-2-ylmethylamino)-9-(phenylmethyl)-8,10-dihydro-7H-[1,2,4]triazolo[3,4-a][2,7]naphthyridine-6-carbonitrile, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
4PLK
 
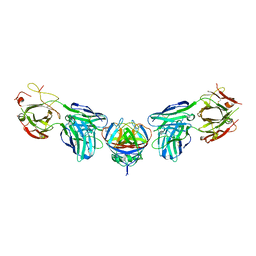 | |
4PLJ
 
 | |
6L10
 
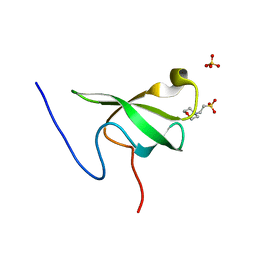 | | PHF20L1 Tudor1 - MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1F
 
 | |
4R7I
 
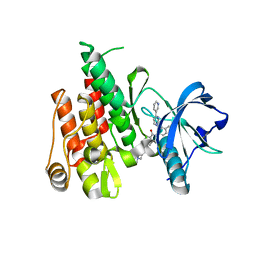 | |
1PZV
 
 | | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans | | Descriptor: | Probable ubiquitin-conjugating enzyme E2-19 kDa | | Authors: | Schormann, N, Lin, G, Li, S, Symersky, J, Qiu, S, Finley, J, Luo, D, Stanton, A, Carson, M, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-07-14 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans
To be Published
|
|
6FWB
 
 | | Crystal structure of Mat2A at 1.79 Angstron resolution | | Descriptor: | GLYCEROL, S-adenosylmethionine synthase isoform type-2, SODIUM ION, ... | | Authors: | Zhou, A, Wei, Z, Bai, J, Wang, H. | | Deposit date: | 2018-03-06 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of a natural inhibitor of methionine adenosyltransferase 2A regulating one-carbon metabolism in keratinocytes.
Ebiomedicine, 39, 2019
|
|
4RT7
 
 | | Crystal Structure of FLT3 with a small molecule inhibitor | | Descriptor: | 1-(5-tert-butyl-1,2-oxazol-3-yl)-3-(4-{7-[2-(morpholin-4-yl)ethoxy]imidazo[2,1-b][1,3]benzothiazol-2-yl}phenyl)urea, Receptor-type tyrosine-protein kinase FLT3 | | Authors: | Zhang, Y, Zhang, C. | | Deposit date: | 2014-11-13 | | Release date: | 2015-04-22 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Characterizing and Overriding the Structural Mechanism of the Quizartinib-Resistant FLT3 "Gatekeeper" F691L Mutation with PLX3397.
Cancer Discov, 5, 2015
|
|
4RC8
 
 | |
4QUW
 
 | |
4RC7
 
 | |
4RC5
 
 | |
3ZXH
 
 | |
3KTZ
 
 | | Structure of GAP31 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome-inactivating protein gelonin | | Authors: | Kong, X.-P. | | Deposit date: | 2009-11-26 | | Release date: | 2010-01-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new activity of anti-HIV and anti-tumor protein GAP31: DNA adenosine glycosidase--structural and modeling insight into its functions.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
3KU0
 
 | | Structure of GAP31 with adenine at its binding pocket | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Ribosome-inactivating protein gelonin | | Authors: | Kong, X.-P. | | Deposit date: | 2009-11-26 | | Release date: | 2010-01-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new activity of anti-HIV and anti-tumor protein GAP31: DNA adenosine glycosidase--structural and modeling insight into its functions.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
5DD6
 
 | |
5DD3
 
 | |
5DD5
 
 | |
6LS4
 
 | | A novel anti-tumor agent S-40 in complex with tubulin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[(4-cyclopropylphenyl)sulfonylamino]-4-methyl-N-(pyridin-3-ylmethyl)benzamide, GLYCEROL, ... | | Authors: | Du, T, Lin, S, Ji, M, Xue, N, Liu, Y, Zhang, K, Lu, D, Chen, X, Xu, H. | | Deposit date: | 2020-01-17 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel orally active microtubule destabilizing agent S-40 targets the colchicine-binding site and shows potent antitumor activity.
Cancer Lett., 495, 2020
|
|
6EAC
 
 | | Pseudomonas syringae SelO | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Tomchick, D.R, Tagliabracci, V.S, Sreelatha, A. | | Deposit date: | 2018-08-02 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Protein AMPylation by an Evolutionarily Conserved Pseudokinase.
Cell, 175, 2018
|
|
6G3E
 
 | | Crystal structure of EDDS lyase in complex with formate | | Descriptor: | Argininosuccinate lyase, FORMIC ACID, SODIUM ION | | Authors: | Poddar, H, Thunnissem, A.M.W.H, Poelarends, G.J. | | Deposit date: | 2018-03-25 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of Ethylenediamine- N, N'-disuccinic Acid Lyase, a Carbon-Nitrogen Bond-Forming Enzyme with a Broad Substrate Scope.
Biochemistry, 57, 2018
|
|
6G3G
 
 | | Crystal structure of EDDS lyase in complex with succinate | | Descriptor: | Argininosuccinate lyase, DI(HYDROXYETHYL)ETHER, SUCCINIC ACID | | Authors: | Poddar, H, Thunnissem, A.M.W.H, Poelarends, G.J. | | Deposit date: | 2018-03-25 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of Ethylenediamine- N, N'-disuccinic Acid Lyase, a Carbon-Nitrogen Bond-Forming Enzyme with a Broad Substrate Scope.
Biochemistry, 57, 2018
|
|
6G3D
 
 | | Crystal structure of Native EDDS lyase | | Descriptor: | Argininosuccinate lyase | | Authors: | Poddar, H, Thunnissem, A.M.W.H, Poelarends, G.J. | | Deposit date: | 2018-03-25 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.221 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of Ethylenediamine- N, N'-disuccinic Acid Lyase, a Carbon-Nitrogen Bond-Forming Enzyme with a Broad Substrate Scope.
Biochemistry, 57, 2018
|
|
5GSA
 
 | | EED in complex with an allosteric PRC2 inhibitor | | Descriptor: | Histone-lysine N-methyltransferase EZH2, N-(furan-2-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of
Nat. Chem. Biol., 13, 2017
|
|
