7EJ7
 
 | | Yeast Dmc1 post-synaptic complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Zhao, L.Y, Xu, J.F, Wang, H.W. | | Deposit date: | 2021-04-01 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Mechanisms of distinctive mismatch tolerance between Rad51 and Dmc1 in homologous recombination.
Nucleic Acids Res., 49, 2021
|
|
7DZH
 
 | | intermediate of FABP with a delay time of 100 ns | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZE
 
 | | Fabp ground state captured by XFELs | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZF
 
 | | Intermediate of FABP with a delay time of 10 ns | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZG
 
 | | Intermediate of FABP with a delay time of 30 ns | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZI
 
 | | intermediate of FABP with a delay time of 300 ns | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZJ
 
 | | Fabp protein before hv | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZK
 
 | | Fabp protein after hv | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZL
 
 | | A69C-M71L mutant of Fabp protein | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
6NW9
 
 | | CRYSTAL STRUCTURE OF A TAILSPIKE PROTEIN 3 (TSP3, ORF212) FROM ESCHERICHIA COLI O157:H7 BACTERIOPHAGE CBA120 | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, CHLORIDE ION, ... | | Authors: | Greenfield, J.Y, Herzberg, O. | | Deposit date: | 2019-02-06 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and tailspike glycosidase machinery of ORF212 from E. coli O157:H7 phage CBA120 (TSP3).
Sci Rep, 9, 2019
|
|
6HHF
 
 | | Crystal Structure of AKT1 in Complex with Covalent-Allosteric AKT Inhibitor Borussertib | | Descriptor: | Borussertib, RAC-alpha serine/threonine-protein kinase | | Authors: | Landel, I, Weisner, J, Mueller, M.P, Scheinpflug, R, Rauh, D. | | Deposit date: | 2018-08-28 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Preclinical Efficacy of Covalent-Allosteric AKT Inhibitor Borussertib in Combination with Trametinib inKRAS-Mutant Pancreatic and Colorectal Cancer.
Cancer Res., 79, 2019
|
|
7BQ9
 
 | | Crystal structure of ASFV p15 | | Descriptor: | 60 kDa polyprotein | | Authors: | Fu, D, Chen, C, Guo, Y. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.612 Å) | | Cite: | Structure of a bifunctional membrane-DNA binding protein, African swine fever virus p15
Protein Cell, 2020
|
|
6L8O
 
 | | Crystal structure of the K. lactis Rad5 (Hg-derivative) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA repair protein RAD5, MERCURY (II) ION | | Authors: | Shen, M, Xiang, S. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Nat Commun, 12, 2021
|
|
6L8N
 
 | | Crystal structure of the K. lactis Rad5 | | Descriptor: | DNA repair protein RAD5, ZINC ION | | Authors: | Shen, M, Xiang, S. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Nat Commun, 12, 2021
|
|
8OTV
 
 | | Crystal structure of NUDT14 complexed with novel compound | | Descriptor: | 1-(1-methylpiperidin-4-yl)-3-(4-phenoxyphenyl)pyrazolo[3,4-d]pyrimidin-4-amine, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Balikci, E, Feyerherm, C, Bradshaw, W, Seupel, R, Brennan, P.E, Bountra, C, von Delft, F, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Unexpected Noncovalent Off-Target Activity of Clinical BTK Inhibitors Leads to Discovery of a Dual NUDT5/14 Antagonist.
J.Med.Chem., 67, 2024
|
|
1C5G
 
 | | PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Descriptor: | PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Authors: | Goldsmith, E.J. | | Deposit date: | 1999-12-07 | | Release date: | 1999-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering of plasminogen activator inhibitor-1 to reduce the rate of latency transition.
Nat.Struct.Biol., 2, 1995
|
|
4WQP
 
 | |
7P3I
 
 | | Crystal structure of human CD40/TNFRSF5 in complex with the anti-CD40 DARPin protein | | Descriptor: | Darpin, SODIUM ION, Tumor necrosis factor receptor superfamily member 5 | | Authors: | Malvezzi, F, Mangold, S, Hospodarsch, T, Reichen, C, Iss, C, Lammens, A, Krapp, S, Domke, C. | | Deposit date: | 2021-07-07 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Multispecific Anti-CD40 DARPin Construct Induces Tumor-Selective CD40 Activation and Tumor Regression.
Cancer Immunol Res, 10, 2022
|
|
4WPF
 
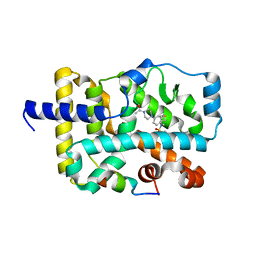 | | Crystal structure of RORc in complex with a phenyl sulfonamide agonist | | Descriptor: | N-[4-(4-acetylpiperazin-1-yl)-2-fluorobenzyl]-N-cyclobutylbenzenesulfonamide, Nuclear receptor ROR-gamma, RHKILHRLLQEGSPS | | Authors: | Kiefer, J.R, Wallweber, H.A, de Leon Boenig, G, Hymowitz, S.G. | | Deposit date: | 2014-10-18 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Minor Structural Change to Tertiary Sulfonamide RORc Ligands Led to Opposite Mechanisms of Action.
Acs Med.Chem.Lett., 6, 2015
|
|
4WLB
 
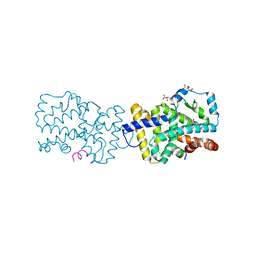 | | Crystal structure of RORc in complex with a partial inverse agonist compound | | Descriptor: | L(+)-TARTARIC ACID, N-(4-fluorobenzyl)-N-(2-methylpropyl)-6-{[1-(methylsulfonyl)piperidin-4-yl]amino}pyridine-3-sulfonamide, Nuclear receptor ROR-gamma, ... | | Authors: | Boenig, G, Hymowitz, S.G, Kiefer, J.R. | | Deposit date: | 2014-10-07 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | A reversed sulfonamide series of selective RORc inverse agonists.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
7QO4
 
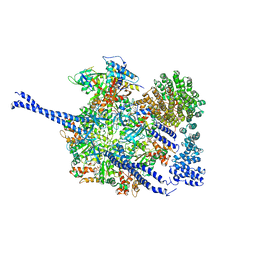 | | 26S proteasome WT-Ubp6-UbVS complex in the si state (ATPases, Rpn1, Ubp6, and UbVS) | | Descriptor: | 26S proteasome regulatory subunit RPN1, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
4LSJ
 
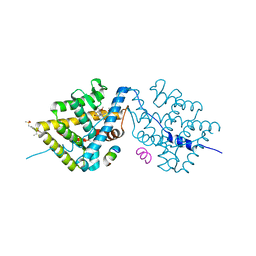 | | Crystal Structure of the Glucocorticoid Receptor Ligand Binding Domain Bound to a Dibenzoxapine Sulfonamide | | Descriptor: | D30 peptide, Glucocorticoid receptor, N-{3-[(1Z)-1-(10-methoxydibenzo[b,e]oxepin-11(6H)-ylidene)propyl]phenyl}methanesulfonamide | | Authors: | Carson, M, Luz, J.G, Clawson, D, Coghlan, M. | | Deposit date: | 2013-07-22 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Glucocorticoid receptor modulators informed by crystallography lead to a new rationale for receptor selectivity, function, and implications for structure-based design.
J.Med.Chem., 57, 2014
|
|
8EKO
 
 | |
1JBU
 
 | | Coagulation Factor VII Zymogen (EGF2/Protease) in Complex with Inhibitory Exosite Peptide A-183 | | Descriptor: | BENZAMIDINE, COAGULATION FACTOR VII, Peptide exosite inhibitor A-183, ... | | Authors: | Eigenbrot, C, Kirchhofer, D, Dennis, M.S, Santell, L, Lazarus, R.A, Stamos, J, Ultsch, M.H. | | Deposit date: | 2001-06-06 | | Release date: | 2001-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The factor VII zymogen structure reveals reregistration of beta strands during activation.
Structure, 9, 2001
|
|
6LKB
 
 | | Crystal Structure of the peptidylprolyl isomerase domain of Arabidopsis thaliana CYP71. | | Descriptor: | COBALT (II) ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Lakhanpal, S, Jobichen, C, Swaminathan, K. | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural and functional analyses of the PPIase domain of Arabidopsis thaliana CYP71 reveal its catalytic activity toward histone H3.
Febs Lett., 595, 2021
|
|
