1H0P
 
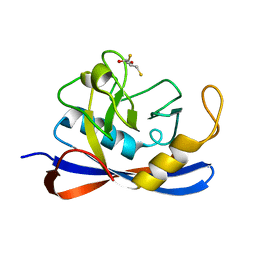 | | Cyclophilin_5 from C. elegans | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE 5 | | Authors: | Picken, N.C, Eschenlauer, S, Taylor, P, Page, A.P, Walkinshaw, M.D. | | Deposit date: | 2002-06-26 | | Release date: | 2002-09-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Biological Characterisation of the Gut-Associated Cyclophilin B Isoforms from Caenorhabditis Elegans
J.Mol.Biol., 322, 2002
|
|
4CHW
 
 | | The electron crystallography structure of the cAMP-free potassium channel MloK1 | | Descriptor: | CYCLIC NUCLEOTIDE-GATED POTASSIUM CHANNEL MLL3241, POTASSIUM ION | | Authors: | Kowal, J, Chami, M, Baumgartner, P, Arheit, M, Chiu, P.L, Rangl, M, Scheuring, S, Schroeder, G.F, Nimigean, C.M, Stahlberg, H. | | Deposit date: | 2013-12-04 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (7 Å) | | Cite: | Ligand-induced structural changes in the cyclic nucleotide-modulated potassium channel MloK1.
Nat Commun, 5, 2014
|
|
4CJ1
 
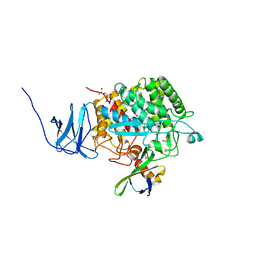 | | Crystal structure of CelD in complex with affitin H3 | | Descriptor: | CALCIUM ION, ENDOGLUCANASE D, GLYCEROL, ... | | Authors: | Correa, A, Pacheco, S, Mechaly, A.E, Obal, G, Behar, G, Mouratou, B, Oppezzo, P, Alzari, P.M, Pecorari, F. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Potent and Specific Inhibition of Glycosidases by Small Artificial Binding Proteins (Affitins)
Plos One, 9, 2014
|
|
4CJ0
 
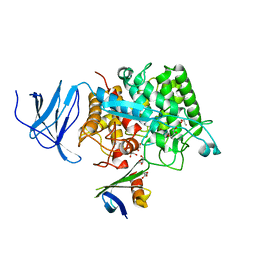 | | Crystal structure of CelD in complex with affitin E12 | | Descriptor: | CALCIUM ION, E12 AFFITIN, ENDOGLUCANASE D, ... | | Authors: | Correa, A, Pacheco, S, Mechaly, A.E, Obal, G, Behar, G, Mouratou, B, Oppezzo, P, Alzari, P.M, Pecorari, F. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Potent and Specific Inhibition of Glycosidases by Small Artificial Binding Proteins (Affitins)
Plos One, 9, 2014
|
|
1G6S
 
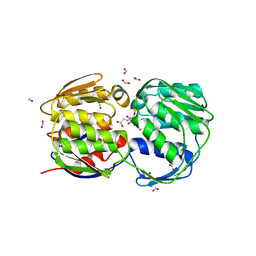 | | STRUCTURE OF EPSP SYNTHASE LIGANDED WITH SHIKIMATE-3-PHOSPHATE AND GLYPHOSATE | | Descriptor: | EPSP SYNTHASE, FORMIC ACID, GLYPHOSATE, ... | | Authors: | Schonbrunn, E, Eschenburg, S, Shuttleworth, W, Schloss, J.V, Amrhein, N, Evans, J.N.S, Kabsch, W. | | Deposit date: | 2000-11-07 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3EMY
 
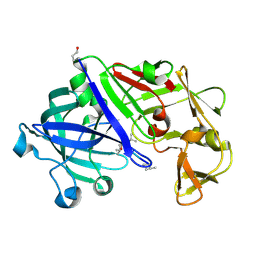 | | Crystal structure of Trichoderma reesei aspartic proteinase complexed with pepstatin A | | Descriptor: | Pepstatin, Trichoderma reesei Aspartic protease | | Authors: | Nascimento, A.S, Krauchenco, S, Golubev, A.M, Gustchina, A, Wlodawer, A, Polikarpov, I. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-07 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Statistical coupling analysis of aspartic proteinases based on crystal
structures of the Trichoderma reesei enzyme and its complex with pepstatin A.
J.Mol.Biol., 382, 2008
|
|
1IG8
 
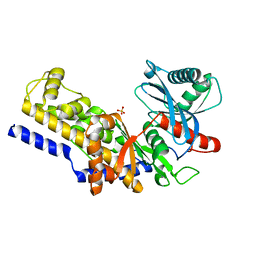 | | Crystal Structure of Yeast Hexokinase PII with the correct amino acid sequence | | Descriptor: | SULFATE ION, hexokinase PII | | Authors: | Kuser, P.R, Krauchenco, S, Antunes, O.A, Polikarpov, I. | | Deposit date: | 2001-04-17 | | Release date: | 2001-05-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The high resolution crystal structure of yeast hexokinase PII with the correct primary sequence provides new insights into its mechanism of action.
J.Biol.Chem., 275, 2000
|
|
5T6R
 
 | | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis: 60S-Nmd3 Complex | | Descriptor: | 25S Ribosomal RNA, 5.8S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Malyutin, A.G, Musalgaonkar, S, Patchett, S, Frank, J, Johnson, A.W. | | Deposit date: | 2016-09-01 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis.
EMBO J., 36, 2017
|
|
2WGP
 
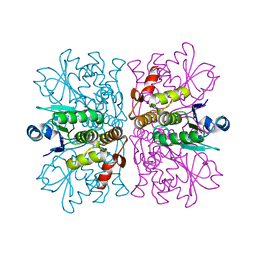 | | Crystal structure of human dual specificity phosphatase 14 | | Descriptor: | DUAL SPECIFICITY PROTEIN PHOSPHATASE 14, PHOSPHATE ION | | Authors: | Lountos, G.T, Tropea, J.E, Cherry, S, Waugh, D.S. | | Deposit date: | 2009-04-22 | | Release date: | 2009-10-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Overproduction, Purification and Structure Determination of Human Dual-Specificity Phosphatase 14.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2V54
 
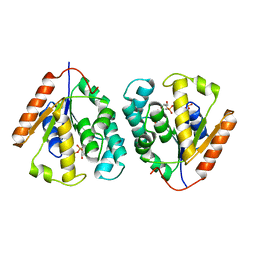 | | Crystal structure of vaccinia virus thymidylate kinase bound to TDP | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE 2-, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Caillat, C, Topalis, D, Agrofoglio, L.A, Pochet, S, Balzarini, J, Deville-Bonne, D, Meyer, P. | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Poxvirus Thymidylate Kinase: An Unexpected Dimerization Has Implications for Antiviral Therapy
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
5T62
 
 | | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis: 60S-Nmd3-Tif6-Lsg1 Complex | | Descriptor: | 25S Ribosomal RNA, 5.8S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Malyutin, A.G, Musalgaonkar, S, Patchett, S, Frank, J, Johnson, A.W. | | Deposit date: | 2016-09-01 | | Release date: | 2017-02-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis.
EMBO J., 36, 2017
|
|
2W0S
 
 | | Crystal structure of vaccinia virus thymidylate kinase bound to brivudin-5'-monophosphate | | Descriptor: | (E)-5-(2-BROMOVINYL)-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Caillat, C, Topalis, D, Agrofoglio, L.A, Pochet, S, Balzarini, J, Deville-Bonne, D, Meyer, P. | | Deposit date: | 2008-10-08 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.918 Å) | | Cite: | Crystal Structure of Poxvirus Thymidylate Kinase: An Unexpected Dimerization Has Implications for Antiviral Therapy
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4NB7
 
 | | Crystal Structure of Two-Domain Laccase from Streptomyces LIvidans AC1709 in complex with azide after 180 min soaking | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, AZIDE ION, ... | | Authors: | Gabdulkhakov, A, Tischenko, S, Yurevich, L, Lisov, A, Leontievsky, A. | | Deposit date: | 2013-10-23 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of Two-Domain Laccase from Streptomyces Lividans AC1709 in complex with azide after 180 min soaking
To be Published
|
|
2VHD
 
 | | Crystal Structure Of The Di-Haem Cytochrome C Peroxidase From Pseudomonas aeruginosa - Mixed Valence Form | | Descriptor: | CALCIUM ION, CYTOCHROME C551 PEROXIDASE, HEME C | | Authors: | Echalier, A, Brittain, T, Wright, J, Boycheva, S, Mortuza, G.B, Fulop, V, Watmough, N.J. | | Deposit date: | 2007-11-20 | | Release date: | 2008-02-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Redox-Linked Structural Changes Associated with the Formation of a Catalytically Competent Form of the Diheme Cytochrome C Peroxidase from Pseudomonas Aeruginosa
Biochemistry, 47, 2008
|
|
4NAJ
 
 | | Crystal Structure of Two-Domain Laccase from Streptomyces Lividans AC1709 in complex with azide after 90 min soaking | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, AZIDE ION, ... | | Authors: | Gabdulkhakov, A, Tischenko, S, Yurevich, L, Lisov, A, Leontievsky, A. | | Deposit date: | 2013-10-22 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Two-Domain Laccase from Streptomyces Lividans AC1709 in complex with azide after 90 min soaking
To be Published
|
|
4N8U
 
 | | Two-Domain Laccase from Streptomyces viridochromogenes at 2.4 A resolution AC629 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Gabdulkhakov, A, Tischenko, S, Yurevich, L, Lisov, A, Leontievsky, A. | | Deposit date: | 2013-10-18 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Two-Domain Laccase from Streptomyces viridochromogenes at 2.4 A resolution AC629
To be Published
|
|
3CTR
 
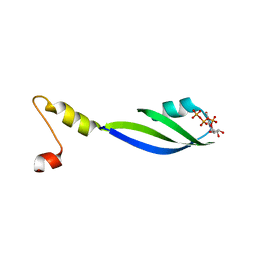 | | Crystal structure of the RRM-domain of the poly(A)-specific ribonuclease PARN bound to m7GTP | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Poly(A)-specific ribonuclease PARN | | Authors: | Monecke, T, Schell, S, Dickmanns, A, Ficner, R. | | Deposit date: | 2008-04-14 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the RRM domain of poly(A)-specific ribonuclease reveals a novel m(7)G-cap-binding mode.
J.Mol.Biol., 382, 2008
|
|
2QX0
 
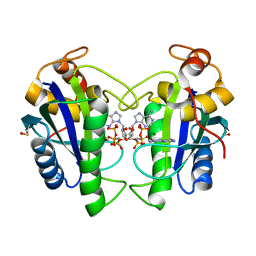 | | Crystal Structure of Yersinia pestis HPPK (Ternary Complex) | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 7,8-dihydro-6-hydroxymethylpterin-pyrophosphokinase, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Blaszczyk, J, Cherry, S, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2007-08-10 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and activity of Yersinia pestis 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase as a novel target for the development of antiplague therapeutics.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5NFA
 
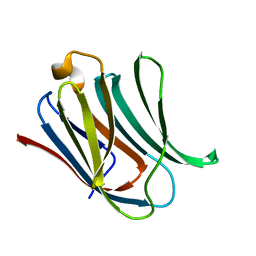 | | Structure of Galectin-3 CRD in complex with compound 3 | | Descriptor: | 3-O-[(3-methoxyphenyl)methyl]-beta-D-galactopyranose-(1-4)-N-acetyl-2-(acetylamino)-2-deoxy-beta-D-glucopyranosylamine, Galectin-3 | | Authors: | Ronin, C, Atmanene, C, Gautier, F.M, Djedaini Pilard, F, Teletchea, S, Ciesielski, F, Vivat Hannah, V, Grandjean, C. | | Deposit date: | 2017-03-13 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Biophysical and structural characterization of mono/di-arylated lactosamine derivatives interaction with human galectin-3.
Biochem. Biophys. Res. Commun., 489, 2017
|
|
5NF9
 
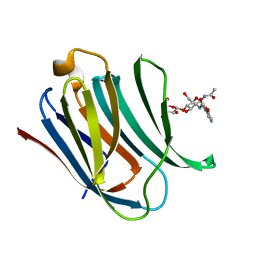 | | Structure of Galectin-3 CRD in complex with compound 2 | | Descriptor: | Galectin-3, ~{N}-[(2~{R},3~{R},4~{R},5~{S},6~{R})-2-acetamido-6-(hydroxymethyl)-5-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4-oxidanyl-oxan-3-yl]-3-methoxy-benzamide | | Authors: | Ronin, C, Atmanene, C, Gautier, F.M, Djedaini Pilard, F, Teletchea, S, Ciesielski, F, Vivat Hannah, V, Grandjean, C. | | Deposit date: | 2017-03-13 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Biophysical and structural characterization of mono/di-arylated lactosamine derivatives interaction with human galectin-3.
Biochem. Biophys. Res. Commun., 489, 2017
|
|
5NFB
 
 | | Structure of Galectin-3 CRD in complex with compound 4 | | Descriptor: | Galectin-3, ~{N}-[(2~{R},3~{R},4~{R},5~{S},6~{R})-2-acetamido-6-(hydroxymethyl)-5-[(2~{S},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-4-[(3-methoxyphenyl)methoxy]-3,5-bis(oxidanyl)oxan-2-yl]oxy-4-oxidanyl-oxan-3-yl]-3-methoxy-benzamide | | Authors: | Ronin, C, Atmanene, C, Gautier, F.M, Djedaini Pilard, F, Teletchea, S, Ciesielski, F, Vivat Hannah, V, Grandjean, C. | | Deposit date: | 2017-03-13 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Biophysical and structural characterization of mono/di-arylated lactosamine derivatives interaction with human galectin-3.
Biochem. Biophys. Res. Commun., 489, 2017
|
|
1K4M
 
 | | Crystal structure of E.coli nicotinic acid mononucleotide adenylyltransferase complexed to deamido-NAD | | Descriptor: | CITRIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NaMN adenylyltransferase | | Authors: | Zhang, H, Zhou, T, Kurnasov, O, Cheek, S, Grishin, N.V, Osterman, A. | | Deposit date: | 2001-10-08 | | Release date: | 2002-10-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of E. coli nicotinate mononucleotide adenylyltransferase and its complex with deamido-NAD.
Structure, 10, 2002
|
|
1K4K
 
 | | Crystal structure of E. coli Nicotinic acid mononucleotide adenylyltransferase | | Descriptor: | Nicotinic acid mononucleotide adenylyltransferase, XENON | | Authors: | Zhang, H, Zhou, T, Kurnasov, O, Cheek, S, Grishin, N.V, Osterman, A.L. | | Deposit date: | 2001-10-08 | | Release date: | 2002-10-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of E. coli nicotinate mononucleotide adenylyltransferase and its complex with deamido-NAD.
Structure, 10, 2002
|
|
5NL2
 
 | | cryo-EM structure of the mTMEM16A ion channel at 6.6 A resolution. | | Descriptor: | Anoctamin-1 | | Authors: | Paulino, C, Neldner, Y, Lam, K.M, Kalienkova, V, Brunner, J.D, Schenck, S, Dutzler, R. | | Deposit date: | 2017-04-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural basis for anion conduction in the calcium-activated chloride channel TMEM16A.
Elife, 6, 2017
|
|
1L3P
 
 | | CRYSTAL STRUCTURE OF THE FUNCTIONAL DOMAIN OF THE MAJOR GRASS POLLEN ALLERGEN Phl p 5b | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, POLLEN ALLERGEN Phl p 5b | | Authors: | Rajashankar, K.R, Bufe, A, Weber, W, Eschenburg, S, Lindner, B, Betzel, C. | | Deposit date: | 2002-02-28 | | Release date: | 2003-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the functional domain of the major grass-pollen allergen Phlp 5b.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
